1Y4Y
 
 | | X-ray crystal structure of Bacillus stearothermophilus Histidine phosphocarrier protein (Hpr) | | Descriptor: | Phosphocarrier protein HPr, SULFATE ION | | Authors: | Sridharan, S, Razvi, A, Scholtz, J.M, Sacchettini, J.C. | | Deposit date: | 2004-12-01 | | Release date: | 2005-02-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The HPr proteins from the thermophile Bacillus stearothermophilus can form domain-swapped dimers.
J.Mol.Biol., 346, 2005
|
|
1Y51
 
 | | X-ray crystal structure of Bacillus stearothermophilus Histidine phosphocarrier protein (Hpr) F29W mutant | | Descriptor: | Phosphocarrier protein HPr, SULFATE ION | | Authors: | Sridharan, S, Razvi, A, Scholtz, J.M, Sacchettini, J.C. | | Deposit date: | 2004-12-01 | | Release date: | 2005-02-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The HPr proteins from the thermophile Bacillus stearothermophilus can form domain-swapped dimers.
J.Mol.Biol., 346, 2005
|
|
2ARO
 
 | | Crystal Structure Of The Native Histone Octamer To 2.1 Angstrom Resolution, Crystalised In The Presence Of S-Nitrosoglutathione | | Descriptor: | CHLORIDE ION, HISTONE H3, HISTONE H4-VI, ... | | Authors: | Wood, C.M, Sodngam, S, Nicholson, J.M, Lambert, S.J, Reynolds, C.D, Baldwin, J.P. | | Deposit date: | 2005-08-20 | | Release date: | 2005-08-30 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The oxidised histone octamer does not form a H3 disulphide bond.
Biochim.Biophys.Acta, 1764, 2006
|
|
4PXT
 
 | |
4PXU
 
 | | Structural basis for the assembly of the mitotic motor kinesin-5 into bipolar tetramers | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Bipolar kinesin KRP-130 | | Authors: | Scholey, J.E, Nithianantham, S, Scholey, J.M, Al-Bassam, J. | | Deposit date: | 2014-03-25 | | Release date: | 2014-04-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.601 Å) | | Cite: | Structural basis for the assembly of the mitotic motor Kinesin-5 into bipolar tetramers.
Elife, 3, 2014
|
|
4OPH
 
 | | X-ray structure of full-length H6N6 NS1 | | Descriptor: | Nonstructural protein 1 | | Authors: | Carrillo, B, Choi, J.M, Bornholdt, Z.A, Sankaran, S, Rice, A.P, Prasad, B.V.V. | | Deposit date: | 2014-02-05 | | Release date: | 2014-02-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.158 Å) | | Cite: | The Influenza A Virus Protein NS1 Displays Structural Polymorphism.
J.Virol., 88, 2014
|
|
4R27
 
 | | Crystal structure of beta-glycosidase BGL167 | | Descriptor: | Glycoside hydrolase | | Authors: | Park, S.J, Choi, J.M, Kyeong, H.H, Kim, S.G, Kim, H.S. | | Deposit date: | 2014-08-09 | | Release date: | 2015-05-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Rational design of a beta-glycosidase with high regiospecificity for triterpenoid tailoring
Chembiochem, 16, 2015
|
|
1QVO
 
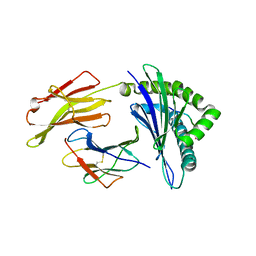 | | STRUCTURES OF HLA-A*1101 IN COMPLEX WITH IMMUNODOMINANT NONAMER AND DECAMER HIV-1 EPITOPES CLEARLY REVEAL THE PRESENCE OF A MIDDLE ANCHOR RESIDUE | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, A-11 alpha chain, ... | | Authors: | Li, L, McNicholl, J.M, Bouvier, M. | | Deposit date: | 2003-08-28 | | Release date: | 2004-06-01 | | Last modified: | 2020-02-19 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Structures of HLA-A*1101 complexed with immunodominant nonamer and decamer HIV-1 epitopes clearly reveal the presence of a middle, secondary anchor residue.
J.Immunol., 172, 2004
|
|
5X6Q
 
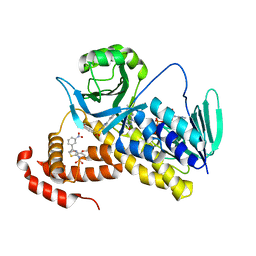 | | Crystal structure of Pseudomonas fluorescens KMO in complex with Ro 61-8048 | | Descriptor: | 3,4-dimethoxy-N-[4-(3-nitrophenyl)-1,3-thiazol-2-yl]benzenesulfonamide, FLAVIN-ADENINE DINUCLEOTIDE, Kynurenine 3-monooxygenase | | Authors: | Kim, H.T, Hwang, K.Y. | | Deposit date: | 2017-02-23 | | Release date: | 2018-02-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.897 Å) | | Cite: | Structural Basis for Inhibitor-Induced Hydrogen Peroxide Production by Kynurenine 3-Monooxygenase
Cell Chem Biol, 25, 2018
|
|
5X6R
 
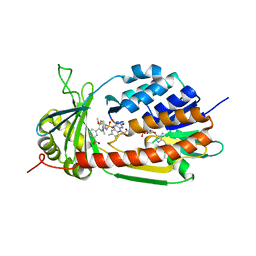 | |
5X68
 
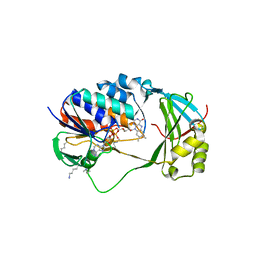 | | Crystal Structure of Human KMO | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Kynurenine 3-monooxygenase | | Authors: | Kim, H.T, Hwang, K.Y. | | Deposit date: | 2017-02-21 | | Release date: | 2018-02-21 | | Last modified: | 2018-05-02 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Basis for Inhibitor-Induced Hydrogen Peroxide Production by Kynurenine 3-Monooxygenase
Cell Chem Biol, 25, 2018
|
|
5X6P
 
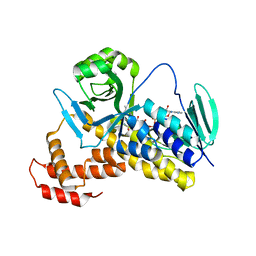 | | Crystal structure of Pseudomonas fluorescens KMO | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Kynurenine 3-monooxygenase | | Authors: | Kim, H.T, Hwang, K.Y. | | Deposit date: | 2017-02-22 | | Release date: | 2018-02-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Structural Basis for Inhibitor-Induced Hydrogen Peroxide Production by Kynurenine 3-Monooxygenase
Cell Chem Biol, 25, 2018
|
|
2ZU6
 
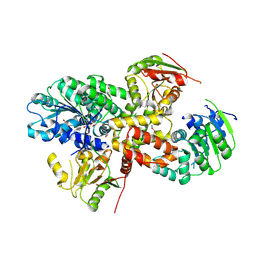 | | crystal structure of the eIF4A-PDCD4 complex | | Descriptor: | 1,2-ETHANEDIOL, ACETIC ACID, Eukaryotic initiation factor 4A-I, ... | | Authors: | Cho, Y, Chang, J.H, Sohn, S.Y. | | Deposit date: | 2008-10-13 | | Release date: | 2009-02-24 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of the eIF4A-PDCD4 complex
Proc.Natl.Acad.Sci.Usa, 106, 2009
|
|
3PJF
 
 | |
3PJE
 
 | |
1FUW
 
 | | SOLUTION STRUCTURE AND BACKBONE DYNAMICS OF A DOUBLE MUTANT SINGLE-CHAIN MONELLIN(SCM) DETERMINED BY NUCLEAR MAGNETIC RESONANCE SPECTROSCOPY | | Descriptor: | MONELLIN | | Authors: | Sung, Y.H, Shin, J, Jung, J, Lee, W. | | Deposit date: | 2000-09-18 | | Release date: | 2001-06-06 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure, backbone dynamics, and stability of a double mutant single-chain monellin. structural origin of sweetness.
J.Biol.Chem., 276, 2001
|
|
1MOL
 
 | |
1V3A
 
 | |
2YFT
 
 | | Crystal structure of inulosucrase from Lactobacillus johnsonii NCC533 in complex with 1-kestose | | Descriptor: | ACETATE ION, CALCIUM ION, LEVANSUCRASE, ... | | Authors: | Pijning, T, Anwar, M.A, Leemhuis, H, Kralj, S, Dijkhuizen, L, Dijkstra, B.W. | | Deposit date: | 2011-04-07 | | Release date: | 2011-08-03 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal Structure of Inulosucrase from Lactobacillus: Insights Into the Substrate Specificity and Product Specificity of Gh68 Fructansucrases.
J.Mol.Biol., 412, 2011
|
|
2YFS
 
 | | Crystal structure of inulosucrase from Lactobacillus johnsonii NCC533 in complex with sucrose | | Descriptor: | CALCIUM ION, LEVANSUCRASE, SULFATE ION, ... | | Authors: | Pijning, T, Anwar, M.A, Leemhuis, H, Kralj, S, Dijkhuizen, L, Dijkstra, B.W. | | Deposit date: | 2011-04-07 | | Release date: | 2011-08-03 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure of Inulosucrase from Lactobacillus: Insights Into the Substrate Specificity and Product Specificity of Gh68 Fructansucrases.
J.Mol.Biol., 412, 2011
|
|
2HJW
 
 | | Crystal Structure of the BC domain of ACC2 | | Descriptor: | Acetyl-CoA carboxylase 2 | | Authors: | Cho, Y.S, Lee, J.I, Shin, D, Kim, H.T, Lee, T.G, Heo, Y.S. | | Deposit date: | 2006-07-02 | | Release date: | 2007-07-03 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of the biotin carboxylase domain of human acetyl-CoA carboxylase 2.
Proteins, 70, 2008
|
|
5L6F
 
 | | Xylooligosaccharide oxidase from Myceliophthora thermophila C1 in complex with Xylobiose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Rozeboom, H.J, Ferrari, A.R, Fraaije, M.W. | | Deposit date: | 2016-05-30 | | Release date: | 2016-09-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Discovery of a Xylooligosaccharide Oxidase from Myceliophthora thermophila C1.
J.Biol.Chem., 291, 2016
|
|
5L6G
 
 | | Xylooligosaccharide oxidase from Myceliophthora thermophila C1 in complex with Xylose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, FAD linked oxidase-like protein, ... | | Authors: | Rozeboom, H.J, Ferrari, A.R, Fraaije, M.W. | | Deposit date: | 2016-05-30 | | Release date: | 2016-09-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Discovery of a Xylooligosaccharide Oxidase from Myceliophthora thermophila C1.
J.Biol.Chem., 291, 2016
|
|
1MNL
 
 | | HIGH-RESOLUTION SOLUTION STRUCTURE OF A SWEET PROTEIN SINGLE-CHAIN MONELLIN (SCM) DETERMINED BY NUCLEAR MAGNETIC RESONANCE SPECTROSCOPY AND DYNAMICAL SIMULATED ANNEALING CALCULATIONS, 21 STRUCTURES | | Descriptor: | MONELLIN | | Authors: | Lee, S.-Y, Lee, J.-H, Chang, H.-J, Jo, J.-M, Jung, J.-W, Lee, W. | | Deposit date: | 1998-08-06 | | Release date: | 1999-06-08 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a sweet protein single-chain monellin determined by nuclear magnetic resonance and dynamical simulated annealing calculations.
Biochemistry, 38, 1999
|
|
1L8O
 
 | | Molecular basis for the local conformational rearrangement of human phosphoserine phosphatase | | Descriptor: | L-3-phosphoserine phosphatase, PHOSPHATE ION, SERINE | | Authors: | Kim, H.Y, Heo, Y.S, Kim, J.H, Park, M.H, Moon, J, Park, S.Y, Lee, T.G, Jeon, Y.H, Ro, S, Hwang, K.Y. | | Deposit date: | 2002-03-21 | | Release date: | 2003-04-01 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Molecular basis for the local conformational rearrangement of human phosphoserine phosphatase
J.Biol.Chem., 277, 2002
|
|
