1N3T
 
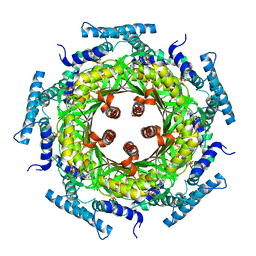 | | Biosynthesis of pteridins. Reaction mechanism of GTP cyclohydrolase I | | Descriptor: | GTP cyclohydrolase I, GUANOSINE-5'-TRIPHOSPHATE | | Authors: | Rebelo, J, Auerbach, G, Bader, G, Bracher, A, Nar, H, Hoesl, C, Schramek, N, Kaiser, J, Bacher, A, Huber, R, Fischer, M. | | Deposit date: | 2002-10-29 | | Release date: | 2003-10-14 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Biosynthesis of Pteridines. Reaction Mechanism of GTP Cyclohydrolase I
J.MOL.BIOL., 326, 2003
|
|
1N3S
 
 | | Biosynthesis of pteridins. Reaction mechanism of GTP cyclohydrolase I | | Descriptor: | GTP cyclohydrolase I, GUANOSINE-5'-TRIPHOSPHATE | | Authors: | Rebelo, J, Auerbach, G, Bader, G, Bracher, A, Nar, H, Hoesl, C, Schramek, N, Kaiser, J, Bacher, A, Huber, R, Fischer, M. | | Deposit date: | 2002-10-29 | | Release date: | 2004-03-02 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Biosynthesis of Pteridines. Reaction Mechanism of GTP Cyclohydrolase I
J.MOL.BIOL., 326, 2003
|
|
1FBX
 
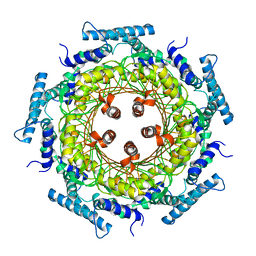 | | CRYSTAL STRUCTURE OF ZINC-CONTAINING E.COLI GTP CYCLOHYDROLASE I | | Descriptor: | CHLORIDE ION, GTP CYCLOHYDROLASE I, ZINC ION | | Authors: | Auerbach, G, Herrmann, A, Bracher, A, Bader, A, Gutlich, M, Fischer, M, Neukamm, M, Nar, H, Garrido-Franco, M, Richardson, J, Huber, R, Bacher, A. | | Deposit date: | 2000-07-17 | | Release date: | 2001-02-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Zinc plays a key role in human and bacterial GTP cyclohydrolase I.
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
1FB1
 
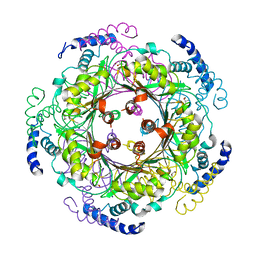 | | CRYSTAL STRUCTURE OF HUMAN GTP CYCLOHYDROLASE I | | Descriptor: | GTP CYCLOHYDROLASE I, ISOPROPYL ALCOHOL, ZINC ION | | Authors: | Auerbach, G, Herrmann, A, Bracher, A, Bader, G, Gutlich, M, Fischer, M, Neukamm, M, Nar, H, Garrido-Franco, M, Richardson, J, Huber, R, Bacher, A. | | Deposit date: | 2000-07-14 | | Release date: | 2000-12-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Zinc plays a key role in human and bacterial GTP cyclohydrolase I.
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
3D2E
 
 | | Crystal structure of a complex of Sse1p and Hsp70, Selenomethionine-labeled crystals | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, GLYCEROL, Heat shock 70 kDa protein 1, ... | | Authors: | Polier, S, Bracher, A. | | Deposit date: | 2008-05-08 | | Release date: | 2008-06-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural basis for the cooperation of Hsp70 and Hsp110 chaperones in protein folding.
Cell(Cambridge,Mass.), 133, 2008
|
|
3D2F
 
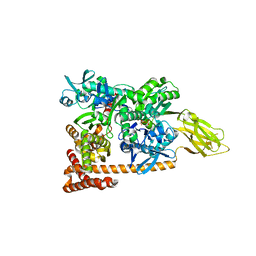 | | Crystal structure of a complex of Sse1p and Hsp70 | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, GLYCEROL, Heat shock 70 kDa protein 1, ... | | Authors: | Polier, S, Bracher, A. | | Deposit date: | 2008-05-08 | | Release date: | 2008-06-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for the cooperation of Hsp70 and Hsp110 chaperones in protein folding.
Cell(Cambridge,Mass.), 133, 2008
|
|
1H2C
 
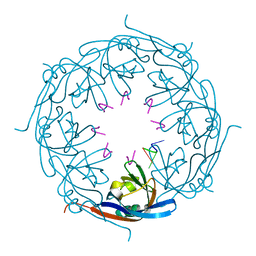 | | Ebola virus matrix protein VP40 N-terminal domain in complex with RNA (High-resolution VP40[55-194] variant). | | Descriptor: | 5'-R(*UP*GP*AP)-3', MATRIX PROTEIN VP40 | | Authors: | Gomis-Ruth, F.X, Dessen, A, Bracher, A, Klenk, H.D, Weissenhorn, W. | | Deposit date: | 2002-08-05 | | Release date: | 2003-04-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The Matrix Protein Vp40 from Ebola Virus Octamerizes Into Pore-Like Structures with Specific RNA Binding Properties
Structure, 11, 2003
|
|
8QXV
 
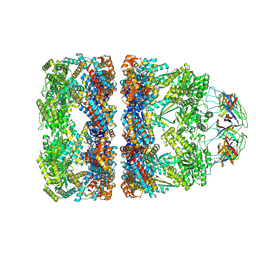 | | In situ structure average of GroEL14-GroES7 complexes with narrow GroEL7 trans ring conformation in Escherichia coli cytosol obtained by cryo electron tomography | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Chaperonin GroEL, ... | | Authors: | Wagner, J, Caravajal, A.I, Beck, F, Bracher, A, Wan, W, Bohn, S, Koerner, R, Baumeister, W, Fernandez-Busnadiego, R, Hartl, F.U. | | Deposit date: | 2023-10-25 | | Release date: | 2024-07-03 | | Last modified: | 2024-09-25 | | Method: | ELECTRON MICROSCOPY (13.6 Å) | | Cite: | Visualizing chaperonin function in situ by cryo-electron tomography.
Nature, 633, 2024
|
|
7AWF
 
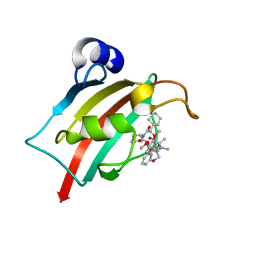 | | The Fk1 domain of FKBP51 in complex with (2R,5S,12R)-12-cyclohexyl-2-[2-(3,4-dimethoxyphenyl)ethyl]-15,15,16-trimethyl-3,19-dioxa-10,13,16-triazatricyclo[18.3.1.0^5,^10]tetracosa-1(24),20,22-triene-4,11,14,17-tetrone | | Descriptor: | (2~{R},5~{S},12~{R})-12-cyclohexyl-2-[2-(3,4-dimethoxyphenyl)ethyl]-15,15,16-trimethyl-3,19-dioxa-10,13,16-triazatricyclo[18.3.1.0^{5,10}]tetracosa-1(24),20,22-triene-4,11,14,17-tetrone, Peptidyl-prolyl cis-trans isomerase FKBP5 | | Authors: | Voll, M.A, Meyners, C, Heymann, T, Merz, S, Purder, P, Bracher, A, Hausch, F. | | Deposit date: | 2020-11-07 | | Release date: | 2021-04-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Macrocyclic FKBP51 Ligands Define a Transient Binding Mode with Enhanced Selectivity.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
7AOU
 
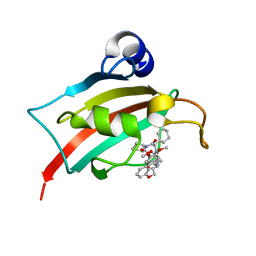 | | The Fk1 domain of FKBP51 in complex with (2'R,5'S,12'R)-12'-cyclohexyl-2'-[2-(3,4-dimethoxyphenyl)ethyl]-3',19'-dioxa-10',13',16'-triazaspiro[cyclopropane-1,15'- tricyclo[18.3.1.0-5,10]tetracosane]-1'(24'),20',22'-triene-4',11',14',17'-tetrone | | Descriptor: | (2'R,5'S,12'R)-12'-cyclohexyl-2'-[2-(3,4-dimethoxyphenyl)ethyl]-3',19'-dioxa-10',13',16'-triazaspiro[cyclopropane-1,15'- tricyclo[18.3.1.0-5,10]tetracosane]-1'(24'),20',22'-triene-4',11',14',17'-tetrone, Peptidyl-prolyl cis-trans isomerase FKBP5 | | Authors: | Voll, M.A, Meyners, C, Heymann, T, Merz, S, Purder, P, Bracher, A, Hausch, F. | | Deposit date: | 2020-10-15 | | Release date: | 2021-04-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.16 Å) | | Cite: | Macrocyclic FKBP51 Ligands Define a Transient Binding Mode with Enhanced Selectivity.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
8R5K
 
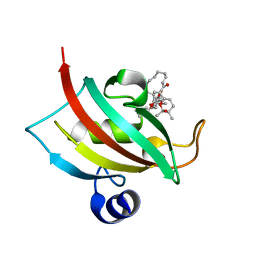 | |
8QXS
 
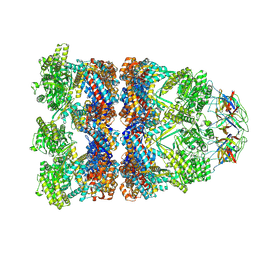 | | CryoEM structure of a GroEL14-GroES7 complex in presence of ADP-BeFx with wide GroEL7 trans ring conformation | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, Chaperonin GroEL, ... | | Authors: | Wagner, J, Caravajal, A.I, Beck, F, Bracher, A, Wan, W, Bohn, S, Koerner, R, Baumeister, W, Fernandez-Busnadiego, R, Hartl, F.U. | | Deposit date: | 2023-10-25 | | Release date: | 2024-07-03 | | Last modified: | 2024-09-25 | | Method: | ELECTRON MICROSCOPY (3.12 Å) | | Cite: | Visualizing chaperonin function in situ by cryo-electron tomography.
Nature, 633, 2024
|
|
8QXU
 
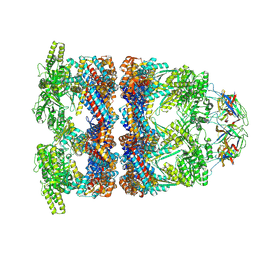 | | In situ structure average of GroEL14-GroES7 complexes with wide GroEL7 trans ring conformation in Escherichia coli cytosol obtained by cryo electron tomography | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Chaperonin GroEL, ... | | Authors: | Wagner, J, Caravajal, A.I, Beck, F, Bracher, A, Wan, W, Bohn, S, Koerner, R, Baumeister, W, Fernandez-Busnadiego, R, Hartl, F.U. | | Deposit date: | 2023-10-25 | | Release date: | 2024-07-03 | | Last modified: | 2024-09-25 | | Method: | ELECTRON MICROSCOPY (12 Å) | | Cite: | Visualizing chaperonin function in situ by cryo-electron tomography.
Nature, 633, 2024
|
|
8QXT
 
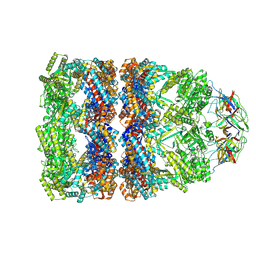 | | CryoEM structure of a GroEL14-GroES7 complex in presence of ADP-BeFx with narrow GroEL7 trans ring conformation | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, Chaperonin GroEL, ... | | Authors: | Wagner, J, Caravajal, A.I, Beck, F, Bracher, A, Wan, W, Bohn, S, Koerner, R, Baumeister, W, Fernandez-Busnadiego, R, Hartl, F.U. | | Deposit date: | 2023-10-25 | | Release date: | 2024-07-03 | | Last modified: | 2024-09-25 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Visualizing chaperonin function in situ by cryo-electron tomography.
Nature, 633, 2024
|
|
4OCN
 
 | |
4N4P
 
 | | Crystal Structure of N-acetylneuraminate lyase from Mycoplasma synoviae, crystal form I | | Descriptor: | Acylneuraminate lyase, CHLORIDE ION | | Authors: | Georgescauld, F, Popova, K, Gupta, A.J, Bracher, A, Engen, J.R, Hayer-Hartl, M, Hartl, F.U. | | Deposit date: | 2013-10-08 | | Release date: | 2014-05-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | GroEL/ES chaperonin modulates the mechanism and accelerates the rate of TIM-barrel domain folding.
Cell(Cambridge,Mass.), 157, 2014
|
|
4N4Q
 
 | | Crystal Structure of N-acetylneuraminate lyase from Mycoplasma synoviae, crystal form II | | Descriptor: | Acylneuraminate lyase | | Authors: | Georgescauld, F, Popova, K, Gupta, A.J, Bracher, A, Engen, J.R, Hayer-Hartl, M, Hartl, F.U. | | Deposit date: | 2013-10-08 | | Release date: | 2014-05-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | GroEL/ES Chaperonin Modulates the Mechanism and Accelerates the Rate of TIM-Barrel Domain Folding.
Cell(Cambridge,Mass.), 157, 2014
|
|
4OCL
 
 | |
4OCM
 
 | |
8P4R
 
 | | In situ structure average of GroEL14-GroES14 complexes in Escherichia coli cytosol obtained by cryo electron tomography | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Chaperonin GroEL, Co-chaperonin GroES, ... | | Authors: | Wagner, J, Caravajal, A.I, Beck, F, Bracher, A, Wan, W, Bohn, S, Koerner, R, Baumeister, W, Fernandez-Busnadiego, R, Hartl, F.U. | | Deposit date: | 2023-05-23 | | Release date: | 2024-07-03 | | Last modified: | 2024-09-25 | | Method: | ELECTRON MICROSCOPY (11.9 Å) | | Cite: | Visualizing chaperonin function in situ by cryo-electron tomography.
Nature, 633, 2024
|
|
8P4M
 
 | | CryoEM structure of a C7-symmetrical GroEL7-GroES7 cage in presence of ADP-BeFx | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, Chaperonin GroEL, ... | | Authors: | Wagner, J, Beck, F, Bracher, A, Caravajal, A.I, Wan, W, Bohn, S, Koerner, R, Baumeister, W, Fernandez-Busnadiego, R, Hartl, F.U. | | Deposit date: | 2023-05-23 | | Release date: | 2024-07-03 | | Last modified: | 2024-09-25 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Visualizing chaperonin function in situ by cryo-electron tomography.
Nature, 633, 2024
|
|
8P4O
 
 | | CryoEM structure of a GroEL7-GroES7 cage with encapsulated ordered substrate MetK in the presence of ADP-BeFx | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, Chaperonin GroEL, ... | | Authors: | Wagner, J, Beck, F, Bracher, A, Caravajal, A.I, Wan, W, Bohn, S, Koerner, R, Baumeister, W, Fernandez-Busnadiego, R, Hartl, F.U. | | Deposit date: | 2023-05-23 | | Release date: | 2024-07-03 | | Last modified: | 2024-09-25 | | Method: | ELECTRON MICROSCOPY (3.04 Å) | | Cite: | Visualizing chaperonin function in situ by cryo-electron tomography.
Nature, 633, 2024
|
|
8P4N
 
 | | CryoEM structure of a GroEL7-GroES7 cage with encapsulated disordered substrate MetK in the presence of ADP-BeFx | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, Chaperonin GroEL, ... | | Authors: | Wagner, J, Beck, F, Bracher, A, Caravajal, A.I, Wan, W, Bohn, S, Koerner, R, Baumeister, W, Fernandez-Busnadiego, R, Hartl, F.U. | | Deposit date: | 2023-05-23 | | Release date: | 2024-07-03 | | Last modified: | 2024-09-25 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Visualizing chaperonin function in situ by cryo-electron tomography.
Nature, 633, 2024
|
|
8P4P
 
 | | Structure average of GroEL14 complexes found in the cytosol of Escherichia coli overexpressing GroEL obtained by cryo electron tomography | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Chaperonin GroEL, ... | | Authors: | Wagner, J, Caravajal, A.I, Beck, F, Bracher, A, Wan, W, Bohn, S, Koerner, R, Baumeister, W, Fernandez-Busnadiego, R, Hartl, F.U. | | Deposit date: | 2023-05-23 | | Release date: | 2024-07-03 | | Last modified: | 2024-09-25 | | Method: | ELECTRON MICROSCOPY (9.6 Å) | | Cite: | Visualizing chaperonin function in situ by cryo-electron tomography.
Nature, 633, 2024
|
|
6EKC
 
 | | Crystal structure of the BSD2 homolog of Arabidopsis thaliana bound to the octameric assembly of RbcL from Thermosynechococcus elongatus | | Descriptor: | DnaJ/Hsp40 cysteine-rich domain superfamily protein, Ribulose bisphosphate carboxylase large chain, ZINC ION | | Authors: | Aigner, H, Wilson, R.H, Bracher, A, Calisse, L, Bhat, J.Y, Hartl, F.U, Hayer-Hartl, M. | | Deposit date: | 2017-09-26 | | Release date: | 2017-12-06 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.63 Å) | | Cite: | Plant RuBisCo assembly in E. coli with five chloroplast chaperones including BSD2.
Science, 358, 2017
|
|
