1OTX
 
 | | Purine Nucleoside Phosphorylase M64V mutant | | Descriptor: | PHOSPHATE ION, Purine nucleoside phosphorylase | | Authors: | Ealick, S.E, Bennett, E.M, Anand, R, Secrist, J.A, Parker, W.B, Hassan, A.E, Allan, P.W, McPherson, D.T, Sorscher, E.J. | | Deposit date: | 2003-03-23 | | Release date: | 2004-02-17 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Designer gene therapy using an Escherichia coli purine nucleoside phosphorylase/prodrug system.
Chem.Biol., 10, 2003
|
|
5FIS
 
 | | Exonuclease domain-containing 1 (Exd1) in the Gd bound conformation | | Descriptor: | EXD1, GADOLINIUM ATOM | | Authors: | Yang, Z, Chen, K.M, Pandey, R.R, Homolka, D, Reuter, M, Rodino Janeiro, B.K, Sachidanandam, R, Fauvarque, M.O, McCarthy, A.A, Pillai, R.S. | | Deposit date: | 2015-10-02 | | Release date: | 2015-12-23 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Piwi Slicing and Exd1 Drive Biogenesis of Nuclear Pirnas from Cytosolic Targets of the Mouse Pirna Pathway
Mol.Cell, 61, 2016
|
|
5FIQ
 
 | | Exonuclease domain-containing 1 (Exd1) in the native conformation | | Descriptor: | EXD1 | | Authors: | Yang, Z, Chen, K.M, Pandey, R.R, Homolka, D, Reuter, M, Rodino Janeiro, B.K, Sachidanandam, R, Fauvarque, M.O, McCarthy, A.A, Pillai, R.S. | | Deposit date: | 2015-10-01 | | Release date: | 2015-12-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Piwi Slicing and Exd1 Drive Biogenesis of Nuclear Pirnas from Cytosolic Targets of the Mouse Pirna Pathway
Mol.Cell, 61, 2016
|
|
6X3N
 
 | | Co-structure of BTK kinase domain with L-005085737 inhibitor | | Descriptor: | 4-[8-azanyl-3-[(2~{S})-1-[4-(dimethylamino)butanoyl]pyrrolidin-2-yl]imidazo[1,5-a]pyrazin-1-yl]-~{N}-(1,3-thiazol-2-yl)benzamide, 4-{8-amino-3-[(6R,8aS)-3-oxohexahydro-3H-[1,3]oxazolo[3,4-a]pyridin-6-yl]imidazo[1,5-a]pyrazin-1-yl}-N-[4-(trifluoromethyl)pyridin-2-yl]benzamide, Tyrosine-protein kinase BTK | | Authors: | Fischmann, T.O. | | Deposit date: | 2020-05-21 | | Release date: | 2020-07-22 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Potent, non-covalent reversible BTK inhibitors with 8-amino-imidazo[1,5-a]pyrazine core featuring 3-position bicyclic ring substitutes.
Bioorg.Med.Chem.Lett., 30, 2020
|
|
6X3O
 
 | | Co-structure of BTK kinase domain with L-005191930 inhibitor | | Descriptor: | 4-[8-azanyl-3-[(2~{S})-1-[4-(dimethylamino)butanoyl]pyrrolidin-2-yl]imidazo[1,5-a]pyrazin-1-yl]-~{N}-(1,3-thiazol-2-yl)benzamide, 4-{8-amino-3-[(6R,8aS)-3-oxo-3,5,6,7,8,8a-hexahydroindolizin-6-yl]imidazo[1,5-a]pyrazin-1-yl}-3-methoxy-N-[4-(trifluoromethyl)pyridin-2-yl]benzamide, Tyrosine-protein kinase BTK | | Authors: | Fischmann, T.O. | | Deposit date: | 2020-05-21 | | Release date: | 2020-07-22 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Potent, non-covalent reversible BTK inhibitors with 8-amino-imidazo[1,5-a]pyrazine core featuring 3-position bicyclic ring substitutes.
Bioorg.Med.Chem.Lett., 30, 2020
|
|
6X3P
 
 | | Co-structure of BTK kinase domain with L-005298385 inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 4-{8-amino-3-[(6R,8aS)-3-oxooctahydroindolizin-6-yl]imidazo[1,5-a]pyrazin-1-yl}-3-(cyclopropyloxy)-N-[4-(trifluoromethyl)pyridin-2-yl]benzamide, ... | | Authors: | Fischmann, T.O. | | Deposit date: | 2020-05-21 | | Release date: | 2020-07-22 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Potent, non-covalent reversible BTK inhibitors with 8-amino-imidazo[1,5-a]pyrazine core featuring 3-position bicyclic ring substitutes.
Bioorg.Med.Chem.Lett., 30, 2020
|
|
8OUY
 
 | | Human RAD51B-RAD51C-RAD51D-XRCC2 (BCDX2) complex, 3.4 A resolution | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, DNA repair protein RAD51 homolog 2, ... | | Authors: | Greenhough, L.A, Liang, C.C, West, S.C. | | Deposit date: | 2023-04-25 | | Release date: | 2023-06-21 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structure and function of the RAD51B-RAD51C-RAD51D-XRCC2 tumour suppressor.
Nature, 619, 2023
|
|
8OUZ
 
 | | Human RAD51B-RAD51C-RAD51D-XRCC2 (BCDX2) complex, 2.2 A resolution | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, DNA repair protein RAD51 homolog 2, ... | | Authors: | Greenhough, L.A, Liang, C.C, West, S.C. | | Deposit date: | 2023-04-25 | | Release date: | 2023-06-21 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (2.2 Å) | | Cite: | Structure and function of the RAD51B-RAD51C-RAD51D-XRCC2 tumour suppressor.
Nature, 619, 2023
|
|
4PXI
 
 | | Elucidation of the Structural and Functional Mechanism of Action of the TetR Family Protein, CprB from S. coelicolor A3(2) | | Descriptor: | CprB, DNA (5'-D(*AP*CP*AP*TP*AP*CP*GP*GP*GP*AP*CP*GP*CP*CP*CP*CP*GP*TP*TP*TP*AP*T)-3'), DNA (5'-D(*AP*TP*AP*AP*AP*CP*GP*GP*GP*GP*CP*GP*TP*CP*CP*CP*GP*TP*AP*TP*GP*T)-3') | | Authors: | Hussain, B, Ruchika, B, Aruna, B, Ruchi, A. | | Deposit date: | 2014-03-24 | | Release date: | 2014-07-02 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural and functional basis of transcriptional regulation by TetR family protein CprB from S. coelicolor A3(2)
Nucleic Acids Res., 42, 2014
|
|
6PCA
 
 | | Crystal structure of beta-ketoadipyl-CoA thiolase | | Descriptor: | ACETATE ION, Beta-ketoadipyl-CoA thiolase, CHLORIDE ION, ... | | Authors: | Sukritee, B, Panjikar, S. | | Deposit date: | 2019-06-17 | | Release date: | 2020-05-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Structural basis for differentiation between two classes of thiolase: Degradative vs biosynthetic thiolase.
J Struct Biol X, 4, 2020
|
|
6PCB
 
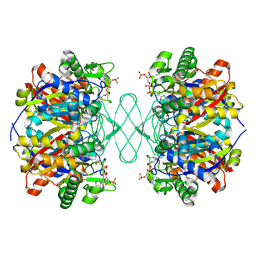 | |
6PCC
 
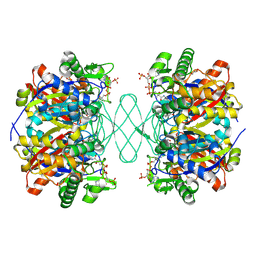 | |
6PCD
 
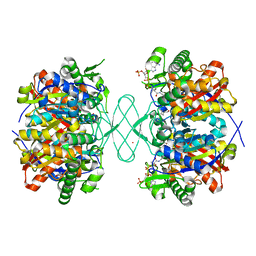 | |
1UW8
 
 | | CRYSTAL STRUCTURE OF OXALATE DECARBOXYLASE | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, MANGANESE (II) ION, OXALATE DECARBOXYLASE OXDC | | Authors: | Just, V.J, Stevenson, C.E.M, Bowater, L, Tanner, A, Lawson, D.M, Bornemann, S. | | Deposit date: | 2004-02-02 | | Release date: | 2004-02-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A Closed Conformation of Bacillus Subtilis Oxalate Decarboxylase Oxdc Provides Evidence for the True Identity of the Active Site
J.Biol.Chem., 279, 2004
|
|
7SVM
 
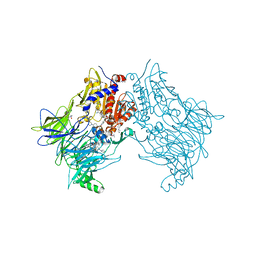 | | DPP8 IN COMPLEX WITH LIGAND ICeD-2 | | Descriptor: | (2S)-2-amino-1-(1,3-dihydro-2H-isoindol-2-yl)-2-[(1r,4S)-4-(pyrrolidin-1-yl)cyclohexyl]ethan-1-one, Dipeptidyl peptidase 8, trimethylamine oxide | | Authors: | Lammens, A, Hollenstein, K, Klein, D.J. | | Deposit date: | 2021-11-19 | | Release date: | 2022-10-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | A Phenotypic Screen Identifies Potent DPP9 Inhibitors Capable of Killing HIV-1 Infected Cells.
Acs Chem.Biol., 17, 2022
|
|
7SVO
 
 | | DPP8 IN COMPLEX WITH LIGAND ICeD-1 | | Descriptor: | (2S,4S)-1-[(2S)-2-amino-2-cyclohexylacetyl]-4-fluoropyrrolidine-2-carbonitrile, Dipeptidyl peptidase 8, trimethylamine oxide | | Authors: | Lammens, A, Hollenstein, K, Klein, D.J. | | Deposit date: | 2021-11-19 | | Release date: | 2022-10-05 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | A Phenotypic Screen Identifies Potent DPP9 Inhibitors Capable of Killing HIV-1 Infected Cells.
Acs Chem.Biol., 17, 2022
|
|
7SVN
 
 | | DPP9 IN COMPLEX WITH LIGAND ICeD-1 | | Descriptor: | (2S,4S)-1-[(2S)-2-amino-2-cyclohexylacetyl]-4-fluoropyrrolidine-2-carbonitrile, Dipeptidyl peptidase 9 | | Authors: | Lammens, A, Hollenstein, K, Klein, D.J. | | Deposit date: | 2021-11-19 | | Release date: | 2022-10-05 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | A Phenotypic Screen Identifies Potent DPP9 Inhibitors Capable of Killing HIV-1 Infected Cells.
Acs Chem.Biol., 17, 2022
|
|
7SVL
 
 | | DPP9 IN COMPLEX WITH LIGAND ICeD-2 | | Descriptor: | (2S)-2-amino-1-(1,3-dihydro-2H-isoindol-2-yl)-2-[(1r,4S)-4-(pyrrolidin-1-yl)cyclohexyl]ethan-1-one, Dipeptidyl peptidase 9 | | Authors: | Lammens, A, Hollenstein, K, Klein, D.J. | | Deposit date: | 2021-11-19 | | Release date: | 2022-10-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | A Phenotypic Screen Identifies Potent DPP9 Inhibitors Capable of Killing HIV-1 Infected Cells.
Acs Chem.Biol., 17, 2022
|
|
2HRY
 
 | | T. maritima PurL complexed with AMPPCP | | Descriptor: | MAGNESIUM ION, PHOSPHATE ION, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, ... | | Authors: | Ealick, S.E, Morar, M. | | Deposit date: | 2006-07-20 | | Release date: | 2007-01-09 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Complexed Structures of Formylglycinamide Ribonucleotide Amidotransferase from Thermotoga maritima Describe a Novel ATP Binding Protein Superfamily
Biochemistry, 45, 2006
|
|
2HS0
 
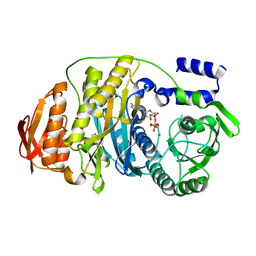 | | T. maritima PurL complexed with ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Phosphoribosylformylglycinamidine synthase II | | Authors: | Ealick, S.E, Morar, M. | | Deposit date: | 2006-07-20 | | Release date: | 2007-01-09 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Complexed Structures of Formylglycinamide Ribonucleotide Amidotransferase from Thermotoga maritima Describe a Novel ATP Binding Protein Superfamily
Biochemistry, 45, 2006
|
|
2HS3
 
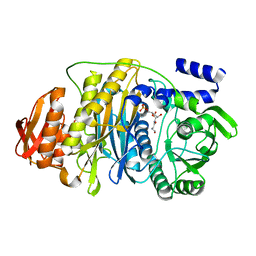 | | T. maritima PurL complexed with FGAR | | Descriptor: | N-(N-FORMYLGLYCYL)-5-O-PHOSPHONO-BETA-D-RIBOFURANOSYLAMINE, PHOSPHATE ION, Phosphoribosylformylglycinamidine synthase II | | Authors: | Ealick, S.E, Morar, M. | | Deposit date: | 2006-07-21 | | Release date: | 2007-01-09 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Complexed Structures of Formylglycinamide Ribonucleotide Amidotransferase from Thermotoga maritima Describe a Novel ATP Binding Protein Superfamily
Biochemistry, 45, 2006
|
|
2HRU
 
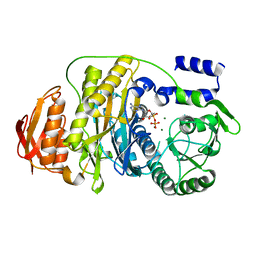 | | T. maritima PurL complexed with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Phosphoribosylformylglycinamidine synthase II | | Authors: | Ealick, S.E, Morar, M. | | Deposit date: | 2006-07-20 | | Release date: | 2007-01-09 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Complexed Structures of Formylglycinamide Ribonucleotide Amidotransferase from Thermotoga maritima Describe a Novel ATP Binding Protein Superfamily
Biochemistry, 45, 2006
|
|
2HS4
 
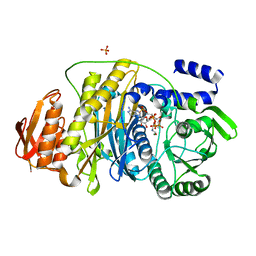 | | T. maritima PurL complexed with FGAR and AMPPCP | | Descriptor: | MAGNESIUM ION, N-(N-FORMYLGLYCYL)-5-O-PHOSPHONO-BETA-D-RIBOFURANOSYLAMINE, PHOSPHATE ION, ... | | Authors: | Ealick, S.E, Morar, M. | | Deposit date: | 2006-07-21 | | Release date: | 2007-01-09 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Complexed Structures of Formylglycinamide Ribonucleotide Amidotransferase from Thermotoga maritima Describe a Novel ATP Binding Protein Superfamily
Biochemistry, 45, 2006
|
|
4D25
 
 | | Crystal structure of the Bombyx mori Vasa helicase (E339Q) in complex with RNA and AMPPNP | | Descriptor: | 5'-R(*UP*GP*AP*CP*AP*UP)-3', BMVLG PROTEIN, GLYCEROL, ... | | Authors: | Spinelli, P, Pillai, R.S, Kadlec, J, Cusack, S. | | Deposit date: | 2014-05-07 | | Release date: | 2014-06-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | RNA Clamping by Vasa Assembles a Pirna Amplifier Complex on Transposon Transcripts.
Cell(Cambridge,Mass.), 157, 2014
|
|
4D26
 
 | | Crystal structure of the Bombyx mori Vasa helicase (E339Q) in complex with RNA,ADP and Pi | | Descriptor: | 5'-R(*UP*GP*AP*CP*AP*UP)-3', ADENOSINE-5'-DIPHOSPHATE, BMVLG PROTEIN, ... | | Authors: | Spinelli, P, Pillai, R.S, Kadlec, J, Cusack, S. | | Deposit date: | 2014-05-07 | | Release date: | 2014-06-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | RNA Clamping by Vasa Assembles a Pirna Amplifier Complex on Transposon Transcripts.
Cell(Cambridge,Mass.), 157, 2014
|
|
