4DRB
 
 | | The crystal structure of FANCM bound MHF complex | | Descriptor: | Centromere protein S, Centromere protein X, Fanconi anemia group M protein | | Authors: | Tao, Y, Niu, L, Teng, M. | | Deposit date: | 2012-02-17 | | Release date: | 2012-05-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.634 Å) | | Cite: | The structure of the FANCM-MHF complex reveals physical features for functional assembly
Nat Commun, 3, 2012
|
|
4EAC
 
 | | Crystal structure of mannonate dehydratase from Escherichia coli strain K12 | | Descriptor: | CHLORIDE ION, MANGANESE (II) ION, Mannonate dehydratase | | Authors: | Qiu, X, Zhu, Y, Yuan, Y, Zhang, Y, Liu, H, Gao, Y, Teng, M, Niu, L. | | Deposit date: | 2012-03-22 | | Release date: | 2013-03-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural insights into decreased enzymatic activity induced by an insert sequence in mannonate dehydratase from Gram negative bacterium.
J.Struct.Biol., 180, 2012
|
|
4ED5
 
 | | Crystal structure of the two N-terminal RRM domains of HuR complexed with RNA | | Descriptor: | 1,2-ETHANEDIOL, 1-METHOXY-2-(2-METHOXYETHOXY)ETHANE, 5'-R(*A*UP*UP*UP*UP*UP*AP*UP*UP*UP*U)-3', ... | | Authors: | Wang, H, Zeng, F, Liu, Q, Niu, L, Teng, M, Li, X. | | Deposit date: | 2012-03-27 | | Release date: | 2012-05-23 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The structure of the ARE-binding domains of Hu antigen R (HuR) undergoes conformational changes during RNA binding.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4E7N
 
 | | Crystal Structure of AhV_TL-I, a Glycosylated Snake-venom Thrombin-like Enzyme from Agkistrodon halys | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, Snake-venom Thrombin-like Enzyme | | Authors: | Zeng, F, Li, X, Teng, M, Niu, L. | | Deposit date: | 2012-03-18 | | Release date: | 2012-04-04 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal Structure of AhV_TL-I, a Glycosylated Snake-venom Thrombin-like Enzyme from Agkistrodon halys
to be published
|
|
4KDC
 
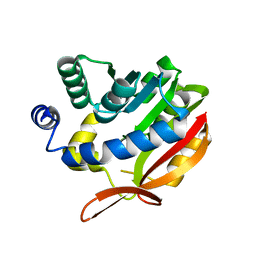 | | Crystal Structure of UBIG | | Descriptor: | 3-demethylubiquinone-9 3-methyltransferase | | Authors: | Zhu, Y, Teng, M, Li, X. | | Deposit date: | 2013-04-24 | | Release date: | 2014-04-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Structural and biochemical studies reveal UbiG/Coq3 as a class of novel membrane-binding proteins.
Biochem. J., 470, 2015
|
|
4F8E
 
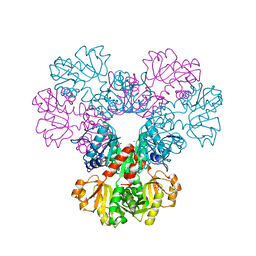 | | Crystal structure of human PRS1 D52H mutant | | Descriptor: | MAGNESIUM ION, Ribose-phosphate pyrophosphokinase 1, SULFATE ION | | Authors: | Chen, P, Teng, M, Li, X. | | Deposit date: | 2012-05-17 | | Release date: | 2013-05-22 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | A small disturbance, but a serious disease: the possible mechanism of D52H-mutant of human PRS1 that causes gout
Iubmb Life, 65, 2013
|
|
4HG9
 
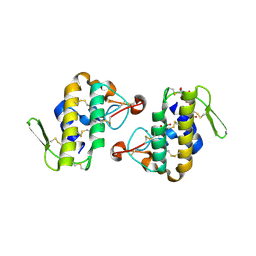 | | Crystal structure of AhV_bPA, a basic PLA2 from Agkistrodon halys pallas venom | | Descriptor: | Basic phospholipase A2 B, CALCIUM ION, CITRIC ACID, ... | | Authors: | Zeng, F, Niu, L, Li, X, Teng, M. | | Deposit date: | 2012-10-07 | | Release date: | 2012-10-17 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of AhV_bPA, a basic PLA2 from Agkistrodon halys pallas venom
TO BE PUBLISHED
|
|
3LBF
 
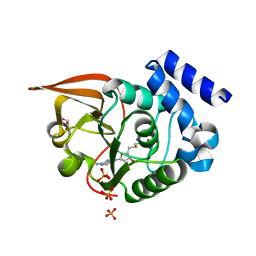 | | Crystal structure of Protein L-isoaspartyl methyltransferase from Escherichia coli | | Descriptor: | GLYCEROL, PHOSPHATE ION, Protein-L-isoaspartate O-methyltransferase, ... | | Authors: | Fang, P, Li, X, Wang, J, Niu, L, Teng, M. | | Deposit date: | 2010-01-08 | | Release date: | 2010-09-08 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of the protein L-isoaspartyl methyltransferase from Escherichia coli
Cell Biochem.Biophys., 58, 2010
|
|
3LKX
 
 | | Human nac dimerization domain | | Descriptor: | Nascent polypeptide-associated complex subunit alpha, Transcription factor BTF3 | | Authors: | Liu, Y, Hu, Y, Li, X, Niu, L, Teng, M. | | Deposit date: | 2010-01-28 | | Release date: | 2010-03-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The crystal structure of the human nascent polypeptide-associated complex domain reveals a nucleic acid-binding region on the NACA subunit
Biochemistry, 49, 2010
|
|
4HG3
 
 | | Structural insights into yeast Nit2: wild-type yeast Nit2 in complex with alpha-ketoglutarate | | Descriptor: | 2-OXOGLUTARIC ACID, CACODYLATE ION, GLYCEROL, ... | | Authors: | Liu, H, Qiu, X, Zhang, M, Gao, Y, Niu, L, Teng, M. | | Deposit date: | 2012-10-06 | | Release date: | 2013-07-31 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Structures of enzyme-intermediate complexes of yeast Nit2: insights into its catalytic mechanism and different substrate specificity compared with mammalian Nit2
Acta Crystallogr.,Sect.D, 69, 2013
|
|
3MD3
 
 | | Crystal Structure of the First Two RRM Domains of Yeast Poly(U) Binding Protein (Pub1) | | Descriptor: | GLYCEROL, Nuclear and cytoplasmic polyadenylated RNA-binding protein PUB1, SULFATE ION | | Authors: | Li, H, Shi, H, Zhu, Z, Wang, H, Niu, L, Teng, M. | | Deposit date: | 2010-03-29 | | Release date: | 2010-05-05 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal Structure of the First Two RRM Domains of Yeast Poly(U) Binding Protein (Pub1)
To be published
|
|
4HGD
 
 | | Structural insights into yeast Nit2: C169S mutant of yeast Nit2 in complex with an endogenous peptide-like ligand | | Descriptor: | CACODYLATE ION, GLYCEROL, N-(4-carboxy-4-oxobutanoyl)-L-cysteinylglycine, ... | | Authors: | Liu, H, Qiu, X, Zhang, M, Gao, Y, Niu, L, Teng, M. | | Deposit date: | 2012-10-08 | | Release date: | 2013-07-31 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Structures of enzyme-intermediate complexes of yeast Nit2: insights into its catalytic mechanism and different substrate specificity compared with mammalian Nit2
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4H5U
 
 | | Structural insights into yeast Nit2: wild-type yeast Nit2 | | Descriptor: | CACODYLATE ION, GLYCEROL, Probable hydrolase NIT2 | | Authors: | Liu, H, Qiu, X, Zhang, M, Gao, Y, Niu, L, Teng, M. | | Deposit date: | 2012-09-18 | | Release date: | 2013-07-31 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Structures of enzyme-intermediate complexes of yeast Nit2: insights into its catalytic mechanism and different substrate specificity compared with mammalian Nit2
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4HG5
 
 | | Structural insights into yeast Nit2: wild-type yeast Nit2 in complex with oxaloacetate | | Descriptor: | CACODYLATE ION, GLYCEROL, OXALOACETATE ION, ... | | Authors: | Liu, H, Qiu, X, Zhang, M, Gao, Y, Niu, L, Teng, M. | | Deposit date: | 2012-10-07 | | Release date: | 2013-07-31 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Structures of enzyme-intermediate complexes of yeast Nit2: insights into its catalytic mechanism and different substrate specificity compared with mammalian Nit2
Acta Crystallogr.,Sect.D, 69, 2013
|
|
3MNM
 
 | | Crystal structure of GAE domain of GGA2p from Saccharomyces cerevisiae | | Descriptor: | ADP-ribosylation factor-binding protein GGA2, GLYCEROL, SULFATE ION | | Authors: | Fang, P, Wang, J, Li, X, Niu, L, Teng, M. | | Deposit date: | 2010-04-21 | | Release date: | 2010-09-08 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Structural basis for the specificity of the GAE domain of yGGA2 for its accessory proteins Ent3 and Ent5
Biochemistry, 49, 2010
|
|
3MIL
 
 | | Crystal structure of isoamyl acetate-hydrolyzing esterase from Saccharomyces cerevisiae | | Descriptor: | GLYCEROL, Isoamyl acetate-hydrolyzing esterase | | Authors: | Ma, J, Lu, Q, Yuan, Y, Li, K, Ge, H, Go, Y, Niu, L, Teng, M. | | Deposit date: | 2010-04-11 | | Release date: | 2010-11-24 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of isoamyl acetate-hydrolyzing esterase from Saccharomyces cerevisiae reveals a novel active site architecture and the basis of substrate specificity
Proteins, 79, 2011
|
|
3MD1
 
 | | Crystal Structure of the Second RRM Domain of Yeast Poly(U)-Binding Protein (Pub1) | | Descriptor: | GLYCEROL, Nuclear and cytoplasmic polyadenylated RNA-binding protein PUB1 | | Authors: | Li, H, Shi, H, Li, Y, Cui, Y, Niu, L, Teng, M. | | Deposit date: | 2010-03-29 | | Release date: | 2010-05-05 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal Structure of the Second RRM Domain of Yeast Poly(U)-Binding Protein (Pub1)
To be published
|
|
3OJB
 
 | |
3P6Y
 
 | | CF Im25-CF Im68-UGUAA complex | | Descriptor: | 5'-R(*UP*GP*UP*AP*A)-3', Cleavage and polyadenylation specificity factor subunit 5, Cleavage and polyadenylation specificity factor subunit 6 | | Authors: | Li, H, Tong, S, Li, X, Shi, H, Gao, Y, Ge, H, Niu, L, Teng, M. | | Deposit date: | 2010-10-11 | | Release date: | 2010-11-03 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis of pre-mRNA recognition by the human cleavage factor Im complex
To be Published
|
|
3P5T
 
 | | CFIm25-CFIm68 complex | | Descriptor: | Cleavage and polyadenylation specificity factor subunit 5, Cleavage and polyadenylation specificity factor subunit 6 | | Authors: | Li, H, Tong, S, Li, X, Shi, H, Gao, Y, Ge, H, Niu, L, Teng, M. | | Deposit date: | 2010-10-11 | | Release date: | 2010-11-03 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis of pre-mRNA recognition by the human cleavage factor Im complex
To be Published
|
|
3R27
 
 | | Crystal structure of the first RRM domain of heterogeneous nuclear ribonucleoprotein L (HnRNP L) | | Descriptor: | GLYCEROL, Heterogeneous nuclear ribonucleoprotein L | | Authors: | Zhang, W, Liu, Y, Zeng, F, Niu, L, Teng, M, Li, X. | | Deposit date: | 2011-03-14 | | Release date: | 2011-09-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Crystal structure of the first RRM domain of heterogeneous nuclear ribonucleoprotein L (HnRNP L)
To be Published
|
|
3S69
 
 | | Crystal structure of saxthrombin | | Descriptor: | CALCIUM ION, Thrombin-like enzyme defibrase | | Authors: | Huang, K, Zhao, W, Teng, M, Niu, L. | | Deposit date: | 2011-05-25 | | Release date: | 2012-05-23 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Structure of saxthrombin, a thrombin-like enzyme from Gloydius saxatilis.
Acta Crystallogr.,Sect.F, 67, 2011
|
|
4H0S
 
 | |
4IQ8
 
 | | Crystal structure of glyceraldehyde-3-phosphate dehydrogenase 3 from Saccharomyces cerevisiae | | Descriptor: | Glyceraldehyde-3-phosphate dehydrogenase 3 | | Authors: | Wang, H, Liu, Q, Niu, L, Teng, M, Li, X. | | Deposit date: | 2013-01-11 | | Release date: | 2013-02-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Preliminary crystallographic analysis of glyceraldehyde-3-phosphate dehydrogenase 3 from Saccharomyces cerevisiae.
Acta Crystallogr.,Sect.F, 68, 2012
|
|
4KR1
 
 | |
