5ZT1
 
 | | Structure of the bacterial pathogens ATPase with substrate ATP gamma S | | Descriptor: | MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, Probable ATP synthase SpaL/MxiB, ... | | Authors: | Gao, X.P, Mu, Z.X, Cui, S. | | Deposit date: | 2018-05-01 | | Release date: | 2018-05-16 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.114 Å) | | Cite: | Structural Insight Into Conformational Changes Induced by ATP Binding in a Type III Secretion-Associated ATPase FromShigella flexneri
Front Microbiol, 9, 2018
|
|
5ZO2
 
 | | Crystal structure of mouse nectin-like molecule 4 (mNecl-4) full ectodomain in complex with mouse nectin-like molecule 1 (mNecl-1) Ig1 domain, 3.3A | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Cell adhesion molecule 3, Cell adhesion molecule 4 | | Authors: | Liu, X, An, T, Li, D, Fan, Z, Xiang, P, Li, C, Ju, W, Li, J, Hu, G, Qin, B, Yin, B, Wojdyla, J.A, Wang, M, Yuan, J, Qiang, B, Shu, P, Cui, S, Peng, X. | | Deposit date: | 2018-04-12 | | Release date: | 2019-01-30 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.29 Å) | | Cite: | Structure of the heterophilic interaction between the nectin-like 4 and nectin-like 1 molecules.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
6J7Q
 
 | | Crystal structure of toxin TglT (unusual type guanylyltransferase-like toxin, Rv1045) mutant S78A from Mycobacterium tuberculosis | | Descriptor: | CALCIUM ION, MAGNESIUM ION, guanylyltransferase-like toxin | | Authors: | Yu, X, Gao, X, Zhu, K, Wojdyla, J.A, Wang, M, Cui, S. | | Deposit date: | 2019-01-18 | | Release date: | 2020-05-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Characterization of a toxin-antitoxin system in Mycobacterium tuberculosis suggests neutralization by phosphorylation as the antitoxicity mechanism.
Commun Biol, 3, 2020
|
|
5Z0W
 
 | |
5ZO1
 
 | | Crystal structure of mouse nectin-like molecule 4 (mNecl-4) full ectodomain (Ig1-Ig3), 2.2A | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Cell adhesion molecule 4, GLYCEROL | | Authors: | Liu, X, An, T, Li, D, Fan, Z, Xiang, P, Li, C, Ju, W, Li, J, Hu, G, Qin, B, Yin, B, Wojdyla, J.A, Wang, M, Yuan, J, Qiang, B, Shu, P, Cui, S, Peng, X. | | Deposit date: | 2018-04-12 | | Release date: | 2019-01-30 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.201 Å) | | Cite: | Structure of the heterophilic interaction between the nectin-like 4 and nectin-like 1 molecules.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
8YG1
 
 | | The Dimer Structure of DSR2 alone | | Descriptor: | SIR2-like domain-containing protein | | Authors: | Gao, X, Zhu, H, Cui, S. | | Deposit date: | 2024-02-26 | | Release date: | 2025-03-05 | | Method: | ELECTRON MICROSCOPY (3.74 Å) | | Cite: | Activation of the bacterial defense-associated sirtuin system.
Commun Biol, 8, 2025
|
|
8YGA
 
 | | The tetramer Structure of DSR2 alone | | Descriptor: | SIR2-like domain-containing protein | | Authors: | Gao, X, Zhu, H, Cui, S. | | Deposit date: | 2024-02-26 | | Release date: | 2025-03-05 | | Method: | ELECTRON MICROSCOPY (4.53 Å) | | Cite: | Activation of the bacterial defense-associated sirtuin system.
Commun Biol, 8, 2025
|
|
8YGN
 
 | |
8YGO
 
 | | The complex by DSR2-CTD-SPR with NAD | | Descriptor: | SIR2-like domain-containing protein, SPR | | Authors: | Gao, X, Zhu, H, Cui, S. | | Deposit date: | 2024-02-26 | | Release date: | 2025-03-05 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.29 Å) | | Cite: | Activation of the bacterial defense-associated sirtuin system.
Commun Biol, 8, 2025
|
|
8YGM
 
 | | The cryo-EM Structure of SPR | | Descriptor: | SPR | | Authors: | Gao, X, Zhu, H, Cui, S. | | Deposit date: | 2024-02-26 | | Release date: | 2025-03-05 | | Method: | ELECTRON MICROSCOPY (3.43 Å) | | Cite: | Activation of the bacterial defense-associated sirtuin system.
Commun Biol, 8, 2025
|
|
8YGP
 
 | |
8YGK
 
 | |
8YGC
 
 | | The Dimer Structure of DSR2-SPR | | Descriptor: | SIR2-like domain-containing protein, SPR | | Authors: | Gao, X, Zhu, H, Cui, S. | | Deposit date: | 2024-02-26 | | Release date: | 2025-03-05 | | Method: | ELECTRON MICROSCOPY (4.03 Å) | | Cite: | Activation of the bacterial defense-associated sirtuin system.
Commun Biol, 8, 2025
|
|
8YGF
 
 | |
8YOY
 
 | | Structure of HKU1A RBD with TMPRSS2 | | Descriptor: | Spike protein S1, Transmembrane protease serine 2 | | Authors: | Gao, X, Cui, S, Ding, W, Shang, K, Zhu, H, Zhu, K. | | Deposit date: | 2024-03-14 | | Release date: | 2024-08-28 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.21 Å) | | Cite: | Structural basis for the interaction between human coronavirus HKU1 spike receptor binding domain and its receptor TMPRSS2.
Cell Discov, 10, 2024
|
|
8YQQ
 
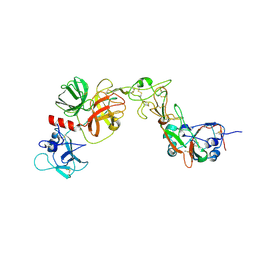 | | Structure of HKU1B RBD with TMPRSS2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1, Transmembrane protease serine 2 | | Authors: | Gao, X, Cui, S, Ding, W, Zhu, K, Shang, K, Zhu, H. | | Deposit date: | 2024-03-19 | | Release date: | 2024-08-28 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.95 Å) | | Cite: | Structural basis for the interaction between human coronavirus HKU1 spike receptor binding domain and its receptor TMPRSS2.
Cell Discov, 10, 2024
|
|
7WYO
 
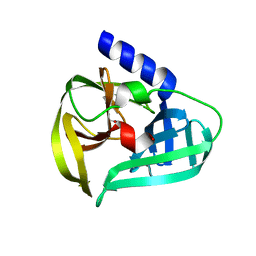 | | Structure of the EV71 3Cpro with 338 inhibitor | | Descriptor: | 3C protein, N-methyl-N-(4,5,6,7-tetrahydro-1,3-benzothiazol-2-ylmethyl)prop-2-enamide | | Authors: | Qin, B, Hou, P, Gao, X, Cui, S. | | Deposit date: | 2022-02-16 | | Release date: | 2022-06-22 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.402 Å) | | Cite: | Acrylamide fragment inhibitors that induce unprecedented conformational distortions in enterovirus 71 3C and SARS-CoV-2 main protease.
Acta Pharm Sin B, 12, 2022
|
|
7WYM
 
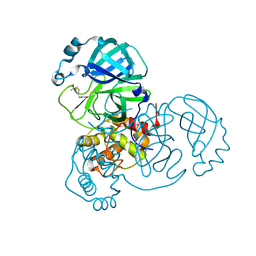 | | Structure of the SARS-COV-2 main protease with 337 inhibitor | | Descriptor: | 3C-like proteinase nsp5, N-methyl-N-[[4-(trifluoromethyl)-1,3-thiazol-2-yl]methyl]prop-2-enamide | | Authors: | Qin, B, Hou, P, Gao, X, Cui, S. | | Deposit date: | 2022-02-16 | | Release date: | 2022-06-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Acrylamide fragment inhibitors that induce unprecedented conformational distortions in enterovirus 71 3C and SARS-CoV-2 main protease.
Acta Pharm Sin B, 12, 2022
|
|
7WYL
 
 | | Structure of the EV71 3Cpro with 337 inhibitor | | Descriptor: | 3C protein, N-methyl-N-[[4-(trifluoromethyl)-1,3-thiazol-2-yl]methyl]prop-2-enamide | | Authors: | Qin, B, Hou, P, Gao, X, Cui, S. | | Deposit date: | 2022-02-16 | | Release date: | 2022-06-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | Acrylamide fragment inhibitors that induce unprecedented conformational distortions in enterovirus 71 3C and SARS-CoV-2 main protease.
Acta Pharm Sin B, 12, 2022
|
|
7WYP
 
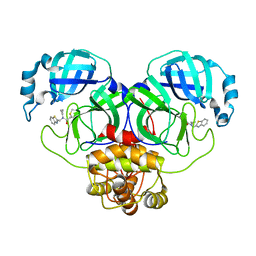 | | Structure of the SARS-COV-2 main protease with EN102 inhibitor | | Descriptor: | 3C-like proteinase, N-(1,3-benzothiazol-2-ylmethyl)-N-cyclopropyl-prop-2-enamide | | Authors: | Qin, B, Hou, P, Gao, X, Cui, S. | | Deposit date: | 2022-02-16 | | Release date: | 2022-06-22 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Acrylamide fragment inhibitors that induce unprecedented conformational distortions in enterovirus 71 3C and SARS-CoV-2 main protease.
Acta Pharm Sin B, 12, 2022
|
|
7CDW
 
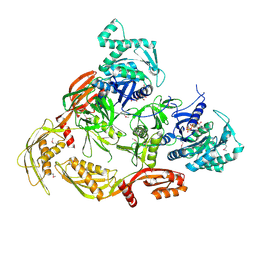 | |
7EK6
 
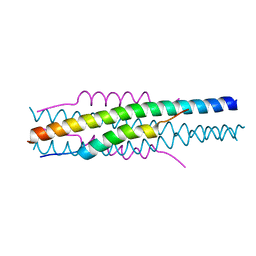 | | Structure of viral peptides IPB19/N52 | | Descriptor: | Spike protein S2 | | Authors: | Yu, D, Qin, B, Cui, S, He, Y. | | Deposit date: | 2021-04-04 | | Release date: | 2021-06-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.243 Å) | | Cite: | Structure-based design and characterization of novel fusion-inhibitory lipopeptides against SARS-CoV-2 and emerging variants.
Emerg Microbes Infect, 10, 2021
|
|
7DHG
 
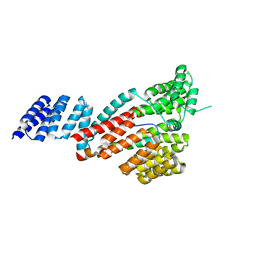 | | Crystal structure of SARS-CoV-2 Orf9b complex with human TOM70 | | Descriptor: | Mitochondrial import receptor subunit TOM70, ORF9b protein | | Authors: | Gao, X, Zhu, K, Qin, B, Olieric, V, Wang, M, Cui, S. | | Deposit date: | 2020-11-14 | | Release date: | 2021-05-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of SARS-CoV-2 Orf9b in complex with human TOM70 suggests unusual virus-host interactions.
Nat Commun, 12, 2021
|
|
7CMD
 
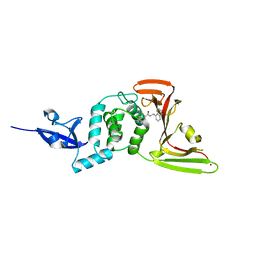 | | Crystal structure of the SARS-CoV-2 PLpro with GRL0617 | | Descriptor: | 5-amino-2-methyl-N-[(1R)-1-naphthalen-1-ylethyl]benzamide, Non-structural protein 3, ZINC ION | | Authors: | Gao, X, Cui, S. | | Deposit date: | 2020-07-27 | | Release date: | 2020-09-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Crystal structure of SARS-CoV-2 papain-like protease.
Acta Pharm Sin B, 11, 2021
|
|
7CJD
 
 | |
