4U7O
 
 | | Active histidine kinase bound with ATP | | Descriptor: | AMP PHOSPHORAMIDATE, Histidine protein kinase sensor protein | | Authors: | Cai, Y, Hu, X, Sang, J. | | Deposit date: | 2014-07-31 | | Release date: | 2015-09-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.395 Å) | | Cite: | Conformational dynamics of the essential sensor histidine kinase WalK.
Acta Crystallogr D Struct Biol, 73, 2017
|
|
1KQ8
 
 | | Solution Structure of Winged Helix Protein HFH-1 | | Descriptor: | HEPATOCYTE NUCLEAR FACTOR 3 FORKHEAD HOMOLOG 1 | | Authors: | Sheng, W, Rance, M, Liao, X. | | Deposit date: | 2002-01-04 | | Release date: | 2002-01-22 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure comparison of two conserved HNF-3/fkh proteins HFH-1 and genesis indicates the existence of folding differences in their complexes with a DNA binding sequence.
Biochemistry, 41, 2002
|
|
4L4M
 
 | | Structural Analysis of a Phosphoribosylated Inhibitor in Complex with Human Nicotinamide Phosphoribosyltransferase | | Descriptor: | 1,2-ETHANEDIOL, N-{4-[(3,5-difluorophenyl)sulfonyl]benzyl}imidazo[1,2-a]pyridine-7-carboxamide, Nicotinamide phosphoribosyltransferase, ... | | Authors: | Oh, A, Ho, Y, Zak, M, Liu, Y, Yuen, P, Zheng, X, Dragovich, S.P, Wang, W. | | Deposit date: | 2013-06-08 | | Release date: | 2014-06-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.445 Å) | | Cite: | Structural and biochemical analyses of the catalysis and potency impact of inhibitor phosphoribosylation by human nicotinamide phosphoribosyltransferase.
Chembiochem, 15, 2014
|
|
8QJR
 
 | | BRG1 bromodomain in complex with VBC via compound 17 | | Descriptor: | (2S,4R)-1-[(2R)-2-[3-[2-[4-[3-[4-[(1R,5S)-3-[3-azanyl-6-(2-hydroxyphenyl)pyridazin-4-yl]-3,8-diazabicyclo[3.2.1]octan-8-yl]pyridin-2-yl]oxycyclobutyl]oxypiperidin-1-yl]ethoxy]-1,2-oxazol-5-yl]-3-methyl-butanoyl]-N-[(1S)-1-[4-(4-methyl-1,3-thiazol-5-yl)phenyl]ethyl]-4-oxidanyl-pyrrolidine-2-carboxamide, CHLORIDE ION, Elongin-B, ... | | Authors: | Kerry, P.S, Hole, A.J, Perez-Dorado, J.I. | | Deposit date: | 2023-09-13 | | Release date: | 2024-01-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.17 Å) | | Cite: | PROTACs Targeting BRM (SMARCA2) Afford Selective In Vivo Degradation over BRG1 (SMARCA4) and Are Active in BRG1 Mutant Xenograft Tumor Models.
J.Med.Chem., 67, 2024
|
|
8QJS
 
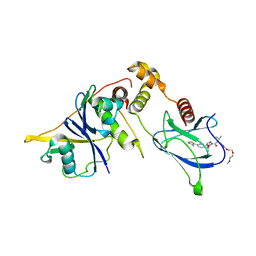 | | VHL/Elongin B/Elongin C complex with compound 155 | | Descriptor: | (2S,4R)-1-[(2R)-2-[3-[2-(2-methoxyethoxy)ethoxy]-1,2-oxazol-5-yl]-3-methyl-butanoyl]-N-[(1S)-1-[4-(4-methyl-1,3-thiazol-5-yl)phenyl]ethyl]-4-oxidanyl-pyrrolidine-2-carboxamide, Elongin-B, Elongin-C, ... | | Authors: | Kerry, P.S, Hole, A.J, Perez-Dorado, J.I. | | Deposit date: | 2023-09-13 | | Release date: | 2024-01-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.191 Å) | | Cite: | PROTACs Targeting BRM (SMARCA2) Afford Selective In Vivo Degradation over BRG1 (SMARCA4) and Are Active in BRG1 Mutant Xenograft Tumor Models.
J.Med.Chem., 67, 2024
|
|
8QJT
 
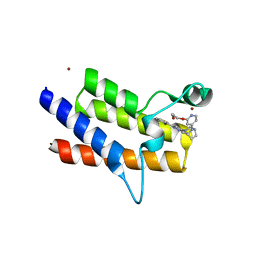 | | BRM (SMARCA2) Bromodomain in complex with ligand 10 | | Descriptor: | 2-[6-azanyl-5-[(1R,5S)-8-[2-(2-methoxyethoxy)pyridin-4-yl]-3,8-diazabicyclo[3.2.1]octan-3-yl]pyridazin-3-yl]phenol, CHLORIDE ION, Probable global transcription activator SNF2L2, ... | | Authors: | Kerry, P.S, Hole, A.J, Perez-Dorado, J.I. | | Deposit date: | 2023-09-13 | | Release date: | 2024-01-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.568 Å) | | Cite: | PROTACs Targeting BRM (SMARCA2) Afford Selective In Vivo Degradation over BRG1 (SMARCA4) and Are Active in BRG1 Mutant Xenograft Tumor Models.
J.Med.Chem., 67, 2024
|
|
7PQY
 
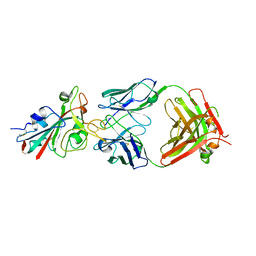 | | Crystal structure of the receptor binding domain of SARS-CoV-2 Spike glycoprotein in complex with FI-3A Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, FI-3A Fab heavy chain, FI-3A Fab light chain, ... | | Authors: | Zhou, D, Ren, J, Stuart, D. | | Deposit date: | 2021-09-20 | | Release date: | 2022-02-02 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structures and therapeutic potential of anti-RBD human monoclonal antibodies against SARS-CoV-2.
Theranostics, 12, 2022
|
|
7PQZ
 
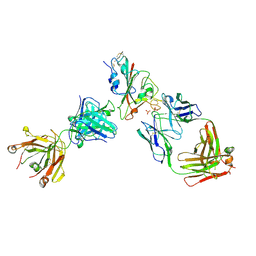 | | Crystal structure of the receptor binding domain of SARS-CoV-2 Spike glycoprotein in complex with FI-3A and FD-11A Fabs | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, FD-11A Fab heavy chain, FD-11A Fab light chain, ... | | Authors: | Zhou, D, Ren, J, Stuart, D.I. | | Deposit date: | 2021-09-20 | | Release date: | 2022-02-02 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structures and therapeutic potential of anti-RBD human monoclonal antibodies against SARS-CoV-2.
Theranostics, 12, 2022
|
|
7PR0
 
 | | Crystal structure of the receptor binding domain of SARS-CoV-2 Spike glycoprotein in complex with FD-5D Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, FD-5D Fab heavy chain, ... | | Authors: | Zhou, D, Ren, J, Stuart, D.I. | | Deposit date: | 2021-09-20 | | Release date: | 2022-02-02 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.92 Å) | | Cite: | Structures and therapeutic potential of anti-RBD human monoclonal antibodies against SARS-CoV-2.
Theranostics, 12, 2022
|
|
3CWG
 
 | | Unphosphorylated mouse STAT3 core fragment | | Descriptor: | Signal transducer and activator of transcription 3 | | Authors: | Ren, Z, Mao, X, Mertens, C, Krishnaraj, R, Qin, J, Mandal, P.K, Romanowshi, M.J, McMurray, J.S. | | Deposit date: | 2008-04-21 | | Release date: | 2008-07-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Crystal structure of unphosphorylated STAT3 core fragment.
Biochem.Biophys.Res.Commun., 374, 2008
|
|
7Q0A
 
 | | SARS-CoV-2 Spike ectodomain with Fab FI3A | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, FI3A fab Light chain, ... | | Authors: | Duyvesteyn, H.M.E, Ren, J, Stuart, D.I. | | Deposit date: | 2021-10-14 | | Release date: | 2022-02-23 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Structures and therapeutic potential of anti-RBD human monoclonal antibodies against SARS-CoV-2.
Theranostics, 12, 2022
|
|
4U7N
 
 | | Inactive structure of histidine kinase | | Descriptor: | Histidine protein kinase sensor protein | | Authors: | Cai, Y, Hu, X, Sang, J. | | Deposit date: | 2014-07-31 | | Release date: | 2015-09-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Conformational dynamics of the essential sensor histidine kinase WalK.
Acta Crystallogr D Struct Biol, 73, 2017
|
|
3SE9
 
 | | Crystal structure of broadly and potently neutralizing antibody VRC-PG04 in complex with HIV-1 gp120 | | Descriptor: | (R,R)-2,3-BUTANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Kwong, P.D, Zhou, T. | | Deposit date: | 2011-06-10 | | Release date: | 2011-08-10 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Focused evolution of HIV-1 neutralizing antibodies revealed by structures and deep sequencing.
Science, 333, 2011
|
|
3SE8
 
 | | Crystal structure of broadly and potently neutralizing antibody VRC03 in complex with HIV-1 gp120 | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Kwong, P.D, Zhou, T. | | Deposit date: | 2011-06-10 | | Release date: | 2011-08-10 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.895 Å) | | Cite: | Focused evolution of HIV-1 neutralizing antibodies revealed by structures and deep sequencing.
Science, 333, 2011
|
|
9INR
 
 | | Crystal structure of PIN1 in complex with inhibitor C3 | | Descriptor: | 3,6,9,12,15,18,21,24,27,30,33,36,39-TRIDECAOXAHENTETRACONTANE-1,41-DIOL, Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1, SULFATE ION, ... | | Authors: | Zhang, L.J, Zhang, L.Y. | | Deposit date: | 2024-07-08 | | Release date: | 2024-09-25 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Re-Evaluating PIN1 as a Therapeutic Target in Oncology Using Neutral Inhibitors and PROTACs.
J.Med.Chem., 67, 2024
|
|
6EFD
 
 | | Hsa Siglec and Unique domains in complex with the sialyl T antigen trisaccharide | | Descriptor: | N-acetyl-alpha-neuraminic acid-(2-3)-beta-D-galactopyranose-(1-3)-2-acetamido-2-deoxy-alpha-D-galactopyranose, SODIUM ION, Streptococcal hemagglutinin | | Authors: | Iverson, T.M. | | Deposit date: | 2018-08-16 | | Release date: | 2020-02-19 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Origins of glycan selectivity in streptococcal Siglec-like adhesins suggest mechanisms of receptor adaptation.
Nat Commun, 13, 2022
|
|
6ER4
 
 | | Ruminococcus gnavus IT-sialidase CBM40 bound to alpha2,6 sialyllactose | | Descriptor: | BNR/Asp-box repeat protein, GLYCEROL, N-acetyl-alpha-neuraminic acid-(2-6)-beta-D-galactopyranose-(1-4)-beta-D-glucopyranose, ... | | Authors: | Owen, C.D, Tailford, L.E, Taylor, G.L, Juge, N. | | Deposit date: | 2017-10-16 | | Release date: | 2018-05-02 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Unravelling the specificity and mechanism of sialic acid recognition by the gut symbiont Ruminococcus gnavus.
Nat Commun, 8, 2017
|
|
6ER2
 
 | |
6EFF
 
 | | NCTC10712 | | Descriptor: | ACETATE ION, CALCIUM ION, GLYCEROL, ... | | Authors: | Iverson, T.M. | | Deposit date: | 2018-08-16 | | Release date: | 2020-02-19 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Origins of glycan selectivity in streptococcal Siglec-like adhesins suggest mechanisms of receptor adaptation.
Nat Commun, 13, 2022
|
|
6FC5
 
 | | Bik1 CAP-Gly domain | | Descriptor: | Microtubule-associated protein | | Authors: | Kumar, A, Stangier, M.M, Steinmetz, M.O. | | Deposit date: | 2017-12-20 | | Release date: | 2018-04-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Structure-Function Relationship of the Bik1-Bim1 Complex.
Structure, 26, 2018
|
|
3CID
 
 | | Structure of BACE Bound to SCH726222 | | Descriptor: | Beta-secretase 1, D(-)-TARTARIC ACID, N'-[(1S,2S)-2-[(4S)-1-benzyl-5-oxoimidazolidin-4-yl]-1-(3,5-difluorobenzyl)-2-hydroxyethyl]-5-methyl-N,N-dipropylbenzene-1,3-dicarboxamide | | Authors: | Strickland, C, Cumming, J. | | Deposit date: | 2008-03-11 | | Release date: | 2008-06-10 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Rational design of novel, potent piperazinone and imidazolidinone BACE1 inhibitors
Bioorg.Med.Chem.Lett., 18, 2008
|
|
3CIC
 
 | | Structure of BACE Bound to SCH709583 | | Descriptor: | Beta-secretase 1, D(-)-TARTARIC ACID, N'-[(1S,2S)-2-[(2S)-4-benzyl-3-oxopiperazin-2-yl]-1-(3,5-difluorobenzyl)-2-hydroxyethyl]-5-methyl-N,N-dipropylbenzene-1,3-dicarboxamide | | Authors: | Strickland, C, Cumming, J. | | Deposit date: | 2008-03-11 | | Release date: | 2008-06-10 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Rational design of novel, potent piperazinone and imidazolidinone BACE1 inhibitors
Bioorg.Med.Chem.Lett., 18, 2008
|
|
2IHK
 
 | |
6ZJA
 
 | | Helicobacter pylori urease with inhibitor bound in the active site | | Descriptor: | 2-{[1-(3,5-dimethylphenyl)-1H-imidazol-2-yl]sulfanyl}-N-hydroxyacetamide, NICKEL (II) ION, Urease subunit alpha, ... | | Authors: | Luecke, H, Cunha, E. | | Deposit date: | 2020-06-28 | | Release date: | 2020-12-23 | | Last modified: | 2025-04-09 | | Method: | ELECTRON MICROSCOPY (2 Å) | | Cite: | Cryo-EM structure of Helicobacter pylori urease with an inhibitor in the active site at 2.0 angstrom resolution.
Nat Commun, 12, 2021
|
|
8D95
 
 | |
