8WRB
 
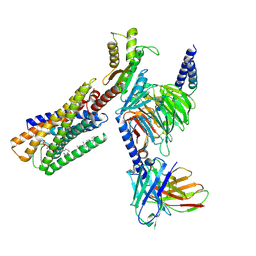 | | Lysophosphatidylserine receptor GPR34-Gi complex | | Descriptor: | Antibody fragment scFv16, CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Gong, W, Liu, G, Li, X, Wang, Y, Zhang, X. | | Deposit date: | 2023-10-13 | | Release date: | 2023-11-08 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (2.91 Å) | | Cite: | Structural basis for ligand recognition and signaling of the lysophosphatidylserine receptors GPR34 and GPR174.
Plos Biol., 21, 2023
|
|
4DKT
 
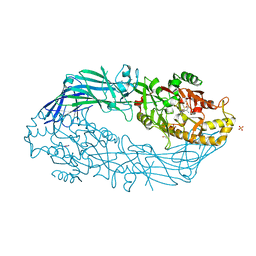 | | Crystal structure of human peptidylarginine deiminase 4 in complex with N-acetyl-L-threonyl-L-alpha-aspartyl-N5-[(1E)-2-fluoroethanimidoyl]-L-ornithinamide | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Protein-arginine deiminase type-4, ... | | Authors: | Jones, J.E, Slack, J.L, Fang, P, Zhang, X, Subramanian, V, Causey, C.P, Coonrod, S.A, Guo, M, Thompson, P.R. | | Deposit date: | 2012-02-04 | | Release date: | 2012-02-29 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.98 Å) | | Cite: | Synthesis and Screening of a Haloacetamidine Containing Library To Identify PAD4 Selective Inhibitors.
Acs Chem.Biol., 7, 2012
|
|
4R2D
 
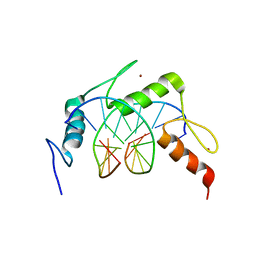 | | Egr1/Zif268 zinc fingers in complex with formylated DNA | | Descriptor: | DNA (5'-D(*AP*GP*CP*GP*TP*GP*GP*GP*(5FC)P*GP*T)-3'), DNA (5'-D(*TP*AP*(5FC)P*GP*CP*CP*CP*AP*CP*GP*C)-3'), Early growth response protein 1, ... | | Authors: | Hashimoto, H, Olanrewaju, Y.O, Zheng, Y, Wilson, G.G, Zhang, X, Cheng, X. | | Deposit date: | 2014-08-11 | | Release date: | 2014-10-08 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.088 Å) | | Cite: | Wilms tumor protein recognizes 5-carboxylcytosine within a specific DNA sequence.
Genes Dev., 28, 2014
|
|
4R2P
 
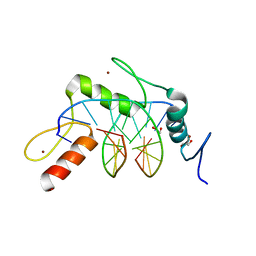 | | Wilms Tumor Protein (WT1) zinc fingers in complex with hydroxymethylated DNA | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*AP*GP*CP*GP*TP*GP*GP*GP*(5HC)P*GP*T)-3'), DNA (5'-D(*TP*AP*(5HC)P*GP*CP*CP*CP*AP*CP*GP*C)-3'), ... | | Authors: | Hashimoto, H, Olanrewaju, Y.O, Zheng, Y, Wilson, G.G, Zhang, X, Cheng, X. | | Deposit date: | 2014-08-12 | | Release date: | 2014-10-08 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.788 Å) | | Cite: | Wilms tumor protein recognizes 5-carboxylcytosine within a specific DNA sequence.
Genes Dev., 28, 2014
|
|
9G13
 
 | | VHH H3-2 in complex with Tau C-terminal peptide | | Descriptor: | Isoform Tau-F of Microtubule-associated protein tau, VHH H3-2 | | Authors: | Dupre, E, Landrieu, I, Danis, C, Hanoulle, X, Mortelecque, J. | | Deposit date: | 2024-07-09 | | Release date: | 2024-07-31 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Inhibition of tau neuronal internalization using anti-tau single domain antibodies.
Nat Commun, 16, 2025
|
|
7ZI4
 
 | | Cryo-EM structure of the human INO80 complex bound to a WT nucleosome | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin-related protein 5, BERYLLIUM TRIFLUORIDE ION, ... | | Authors: | Vance, N.R, Ayala, R, Willhoft, O, Tvardovskiy, A, McCormack, E.A, Bartke, T, Zhang, X, Wigley, D.B. | | Deposit date: | 2022-04-07 | | Release date: | 2023-04-19 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM structure of the human INO80 complex bound to a WT nucleosome
To Be Published
|
|
3GT8
 
 | | Crystal structure of the inactive EGFR kinase domain in complex with AMP-PNP | | Descriptor: | Epidermal growth factor receptor, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Jura, N, Endres, N.F, Engel, K, Deindl, S, Das, R, Lamers, M.H, Wemmer, D.E, Zhang, X, Kuriyan, J. | | Deposit date: | 2009-03-27 | | Release date: | 2009-07-21 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.955 Å) | | Cite: | Mechanism for activation of the EGF receptor catalytic domain by the juxtamembrane segment.
Cell(Cambridge,Mass.), 137, 2009
|
|
4F5X
 
 | | Location of the dsRNA-dependent polymerase, VP1, in rotavirus particles | | Descriptor: | Intermediate capsid protein VP6, RNA-directed RNA polymerase, VP2 protein, ... | | Authors: | Estrozi, L.F, Settembre, E.C, Goret, G, McClain, B, Zhang, X, Chen, J.Z, Grigorieff, N, Harrison, S.C. | | Deposit date: | 2012-05-13 | | Release date: | 2012-10-24 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (5 Å) | | Cite: | Location of the dsRNA-Dependent Polymerase, VP1, in Rotavirus Particles.
J.Mol.Biol., 425, 2013
|
|
4DHZ
 
 | | The structure of h/ceOTUB1-ubiquitin aldehyde-UBC13~Ub | | Descriptor: | Ubiquitin, Ubiquitin aldehyde, Ubiquitin thioesterase otubain-like, ... | | Authors: | Wiener, R, Zhang, X, Wang, T, Wolberger, C. | | Deposit date: | 2012-01-30 | | Release date: | 2012-02-22 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (3.11 Å) | | Cite: | The mechanism of OTUB1-mediated inhibition of ubiquitination.
Nature, 483, 2012
|
|
8WQL
 
 | | In situ PBS-PSII supercomplex from cyanobacterial Spirulina platensis | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | You, X, Zhang, X, Xiao, Y.N, Sun, S, Sui, S.F. | | Deposit date: | 2023-10-11 | | Release date: | 2024-07-31 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structure of in situ PBS-PSII supercomplex at 3.5 Angstroms resolution.
To Be Published
|
|
4C6R
 
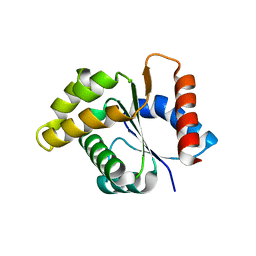 | | Crystal structure of the TIR domain from the Arabidopsis Thaliana disease resistance protein RPS4 | | Descriptor: | DISEASE RESISTANCE PROTEIN RPS4 | | Authors: | Williams, S.J, Sohn, K.H, Wan, L, Bernoux, M, Ma, Y, Segonzac, C, Ve, T, Sarris, P, Ericsson, D.J, Saucet, S.B, Zhang, X, Parker, J, Dodds, P.N, Jones, J.D.G, Kobe, B. | | Deposit date: | 2013-09-19 | | Release date: | 2014-05-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural Basis for Assembly and Function of a Heterodimeric Plant Immune Receptor.
Science, 344, 2014
|
|
4C6S
 
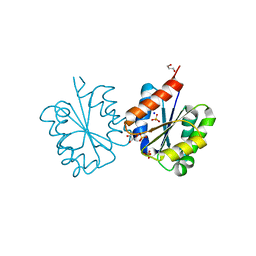 | | Crystal structure of the TIR domain from the Arabidopsis Thaliana disease resistance protein RRS1 | | Descriptor: | GLYCEROL, PROBABLE WRKY TRANSCRIPTION FACTOR 52, SODIUM ION, ... | | Authors: | Wan, L, Williams, S.J, Sohn, K.H, Bernoux, M, Ma, Y, Segonzac, C, Ve, T, Sarris, P, Ericsson, D.J, Saucet, S.B, Zhang, X, Parker, J, Dodds, P.N, Jones, J.D.G, Kobe, B. | | Deposit date: | 2013-09-19 | | Release date: | 2014-05-28 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.751 Å) | | Cite: | Structural Basis for Assembly and Function of a Heterodimeric Plant Immune Receptor.
Science, 344, 2014
|
|
4R2S
 
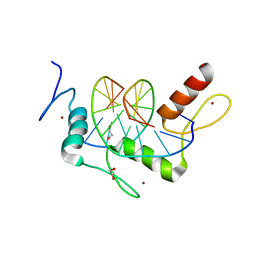 | | Wilms Tumor Protein (WT1) Q369P zinc fingers in complex with methylated DNA | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*AP*GP*CP*GP*TP*GP*GP*GP*(5CM)P*GP*T)-3'), DNA (5'-D(*TP*AP*(5CM)P*GP*CP*CP*CP*AP*CP*GP*C)-3'), ... | | Authors: | Hashimoto, H, Olanrewaju, Y.O, Zheng, Y, Wilson, G.G, Zhang, X, Cheng, X. | | Deposit date: | 2014-08-12 | | Release date: | 2014-10-08 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.489 Å) | | Cite: | Wilms tumor protein recognizes 5-carboxylcytosine within a specific DNA sequence.
Genes Dev., 28, 2014
|
|
3JB5
 
 | |
2UXP
 
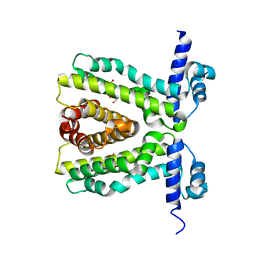 | | TtgR in complex Chloramphenicol | | Descriptor: | CHLORAMPHENICOL, HTH-TYPE TRANSCRIPTIONAL REGULATOR TTGR | | Authors: | Alguel, Y, Meng, C, Teran, W, Krell, T, Ramos, J.L, Gallegos, M.-T, Zhang, X. | | Deposit date: | 2007-03-29 | | Release date: | 2007-05-08 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal Structures of Multidrug Binding Protein Ttgr in Complex with Antibiotics and Plant Antimicrobials.
J.Mol.Biol., 369, 2007
|
|
2UXH
 
 | | TtgR in complex with Quercetin | | Descriptor: | 3,5,7,3',4'-PENTAHYDROXYFLAVONE, HTH-TYPE TRANSCRIPTIONAL REGULATOR TTGR | | Authors: | Alguel, Y, Meng, C, Teran, W, Krell, T, Ramos, J.L, Gallegos, M.-T, Zhang, X. | | Deposit date: | 2007-03-28 | | Release date: | 2007-05-08 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structures of Multidrug Binding Protein Ttgr in Complex with Antibiotics and Plant Antimicrobials.
J.Mol.Biol., 369, 2007
|
|
8IZB
 
 | | Lysophosphatidylserine receptor GPR174-Gs complex | | Descriptor: | CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Gong, W, Liu, G, Li, X, Zhang, X. | | Deposit date: | 2023-04-06 | | Release date: | 2023-11-01 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.06 Å) | | Cite: | Structural basis for ligand recognition and signaling of the lysophosphatidylserine receptors GPR34 and GPR174.
Plos Biol., 21, 2023
|
|
2DHR
 
 | | Whole cytosolic region of ATP-dependent metalloprotease FtsH (G399L) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, FtsH | | Authors: | Suno, R, Niwa, H, Tsuchiya, D, Zhang, X, Yoshida, M, Morikawa, K. | | Deposit date: | 2006-03-24 | | Release date: | 2006-06-27 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (3.9 Å) | | Cite: | Structure of the Whole Cytosolic Region of ATP-Dependent Protease FtsH
Mol.Cell, 22, 2006
|
|
1RUN
 
 | | CATABOLITE GENE ACTIVATOR PROTEIN (CAP)/DNA COMPLEX + ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE | | Descriptor: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, DNA (5'-D(*CP*TP*AP*GP*AP*TP*CP*AP*CP*AP*TP*TP*TP*TP*TP*CP*G )-3'), DNA (5'-D(*GP*CP*GP*AP*AP*AP*AP*AP*TP*GP*TP*GP*AP*T)-3'), ... | | Authors: | Parkinson, G.N, Gunasekera, A, Vojtechovsky, J, Zhang, X, Kunkel, T.A, Berman, H.M, Ebright, R.H. | | Deposit date: | 1996-05-26 | | Release date: | 1997-01-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Aromatic hydrogen bond in sequence-specific protein DNA recognition.
Nat.Struct.Biol., 3, 1996
|
|
8IZ4
 
 | | Lysophosphatidylserine receptor GPR34-Gi complex | | Descriptor: | Antibody fragment scFv16, CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Gong, W, Liu, G, Li, X, Zhang, X. | | Deposit date: | 2023-04-06 | | Release date: | 2024-04-10 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (2.93 Å) | | Cite: | Structural mechanisms of ligand binding and signaling in lysophosphatidylserine receptors
To Be Published
|
|
8F4L
 
 | |
2DI4
 
 | | Crystal structure of the FtsH protease domain | | Descriptor: | Cell division protein ftsH homolog, MERCURY (II) ION | | Authors: | Suno, R, Niwa, H, Tsuchiya, D, Zhang, X, Yoshida, M, Morikawa, K. | | Deposit date: | 2006-03-28 | | Release date: | 2006-06-27 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Structure of the Whole Cytosolic Region of ATP-Dependent Protease FtsH
Mol.Cell, 22, 2006
|
|
2B36
 
 | | Crystal structure of Mycobacterium tuberculosis enoyl reductase (InhA) inhibited by 5-pentyl-2-phenoxyphenol | | Descriptor: | 5-PENTYL-2-PHENOXYPHENOL, Enoyl-[acyl-carrier-protein] reductase [NADH], NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Sullivan, T.J, Truglio, J.J, Novichenok, P, Stratton, C, Zhang, X, Kaur, T, Johnson, F, Boyne, M.S, Amin, A. | | Deposit date: | 2005-09-19 | | Release date: | 2006-03-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | High Affinity InhA Inhibitors with Activity against Drug-Resistant Strains
of Mycobacterium tuberculosis
ACS Chem.Biol., 1, 2006
|
|
8JNN
 
 | | linker protein LPP1 from red algal Porphyridium purpureum. | | Descriptor: | LPP1 | | Authors: | You, X, Zhang, X, Cheng, J, Xiao, Y.N, Sun, S, Sui, S.F. | | Deposit date: | 2023-06-06 | | Release date: | 2023-08-02 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structure of lateral hexamer of PBS-PSII-PSI-LHCs megacomplex at 6.3 Angstroms resolution.
To Be Published
|
|
8JNG
 
 | | Methanesulfonate monooxygenase ferredoxin subunit of PBS-PSII-PSI-LHCs from Porphyridium purpureum. | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, Methanesulfonate monooxygenase ferredoxin subunit | | Authors: | You, X, Zhang, X, Cheng, J, Xiao, Y.N, Sun, S, Sui, S.F. | | Deposit date: | 2023-06-06 | | Release date: | 2023-08-02 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structure of lateral hexamer of PBS-PSII-PSI-LHCs megacomplex at 6.3 Angstroms resolution.
To Be Published
|
|
