7ZRD
 
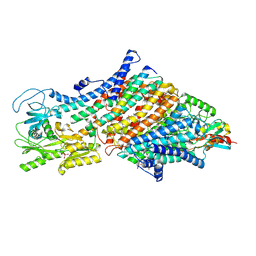 | | Cryo-EM map of the WT KdpFABC complex in the E1-P tight conformation, stabilised with the inhibitor orthovanadate | | Descriptor: | CARDIOLIPIN, POTASSIUM ION, Potassium-transporting ATPase ATP-binding subunit, ... | | Authors: | Hielkema, L, Stock, C, Silberberg, J.M, Corey, R.A, Wunnicke, D, Stansfeld, P.J, Haenelt, I, Paulino, C. | | Deposit date: | 2022-05-04 | | Release date: | 2022-11-16 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Inhibited KdpFABC transitions into an E1 off-cycle state.
Elife, 11, 2022
|
|
7ZRE
 
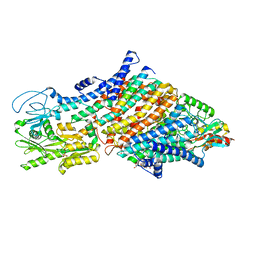 | | Cryo-EM map of the WT KdpFABC complex in the E1-P tight conformation, under turnover conditions | | Descriptor: | CARDIOLIPIN, POTASSIUM ION, Potassium-transporting ATPase ATP-binding subunit, ... | | Authors: | Hielkema, L, Stock, C, Silberberg, J.M, Corey, R.A, Wunnicke, D, Dubach, V.R.A, Stansfeld, P.J, Haenelt, I, Paulino, C. | | Deposit date: | 2022-05-04 | | Release date: | 2022-11-16 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Inhibited KdpFABC transitions into an E1 off-cycle state.
Elife, 11, 2022
|
|
7ZRL
 
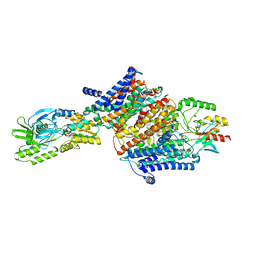 | | Cryo-EM map of the unphosphorylated KdpFABC complex in the E2-P conformation, under turnover conditions | | Descriptor: | POTASSIUM ION, Potassium-transporting ATPase ATP-binding subunit, Potassium-transporting ATPase KdpC subunit, ... | | Authors: | Hielkema, L, Stock, C, Silberberg, J.M, Corey, R.A, Wunnicke, D, Dubach, V.R.A, Stansfeld, P.J, Haenelt, I, Paulino, C. | | Deposit date: | 2022-05-04 | | Release date: | 2022-11-16 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Inhibited KdpFABC transitions into an E1 off-cycle state.
Elife, 11, 2022
|
|
7ZRK
 
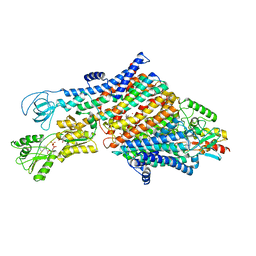 | | Cryo-EM map of the WT KdpFABC complex in the E1-P_ADP conformation, under turnover conditions | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CARDIOLIPIN, POTASSIUM ION, ... | | Authors: | Hielkema, L, Stock, C, Silberberg, J.M, Corey, R.A, Wunnicke, D, Dubach, V.R.A, Stansfeld, P.J, Haenelt, I, Paulino, C. | | Deposit date: | 2022-05-04 | | Release date: | 2022-11-16 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Inhibited KdpFABC transitions into an E1 off-cycle state.
Elife, 11, 2022
|
|
7ZSG
 
 | | Structure of Orange Carotenoid Protein with canthaxanthin bound after 1 minute of illumination | | Descriptor: | ACETATE ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Chukhutsina, V.U, Baxter, J.M, Fadini, A, Morgan, R.M, Pope, M.A, Maghlaoui, K, Orr, C, Wagner, A, van Thor, J.J. | | Deposit date: | 2022-05-06 | | Release date: | 2022-11-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Light activation of Orange Carotenoid Protein reveals bicycle-pedal single-bond isomerization.
Nat Commun, 13, 2022
|
|
7ZRG
 
 | | Cryo-EM map of the WT KdpFABC complex in the E1_ATPearly conformation, under turnover conditions | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CARDIOLIPIN, POTASSIUM ION, ... | | Authors: | Hielkema, L, Stock, C, Silberberg, J.M, Corey, R.A, Rheinberger, J, Wunnicke, D, Dubach, V.R.A, Stansfeld, P.J, Haenelt, I, Paulino, C. | | Deposit date: | 2022-05-04 | | Release date: | 2022-11-16 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Inhibited KdpFABC transitions into an E1 off-cycle state.
Elife, 11, 2022
|
|
7ZRH
 
 | | Cryo-EM structure of the KdpFABC complex in a nucleotide-free E1 conformation loaded with K+ | | Descriptor: | CARDIOLIPIN, POTASSIUM ION, Potassium-transporting ATPase ATP-binding subunit, ... | | Authors: | Hielkema, L, Stock, C, Silberberg, J.M, Corey, R.A, Wunnicke, D, Stansfeld, P.J, Haenelt, I, Paulino, C. | | Deposit date: | 2022-05-04 | | Release date: | 2022-11-16 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Inhibited KdpFABC transitions into an E1 off-cycle state.
Elife, 11, 2022
|
|
7ZRI
 
 | | Cryo-EM structure of the KdpFABC complex in a nucleotide-free E1 conformation loaded with K+ | | Descriptor: | CARDIOLIPIN, POTASSIUM ION, Potassium-transporting ATPase ATP-binding subunit, ... | | Authors: | Hielkema, L, Stock, C, Silberberg, J.M, Corey, R.A, Wunnicke, D, Stansfeld, P.J, Haenelt, I, Paulino, C. | | Deposit date: | 2022-05-04 | | Release date: | 2022-11-16 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Inhibited KdpFABC transitions into an E1 off-cycle state.
Elife, 11, 2022
|
|
7ZRJ
 
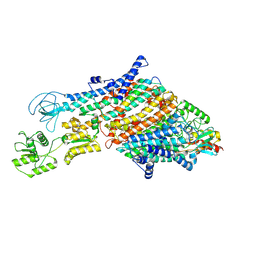 | | Cryo-EM structure of the KdpFABC complex in a nucleotide-free E1 conformation loaded with K+ | | Descriptor: | CARDIOLIPIN, POTASSIUM ION, Potassium-transporting ATPase ATP-binding subunit, ... | | Authors: | Hielkema, L, Stock, C, Silberberg, J.M, Corey, R.A, Wunnicke, D, Stansfeld, P.J, Haenelt, I, Paulino, C. | | Deposit date: | 2022-05-04 | | Release date: | 2022-11-16 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Inhibited KdpFABC transitions into an E1 off-cycle state.
Elife, 11, 2022
|
|
8B4H
 
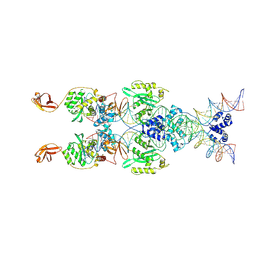 | | IstA transposase cleaved donor complex | | Descriptor: | DNA (55-MER) / right IS21 transposon end (insertion sequence IS5376), DNA (57-MER) / right IS21 transposon end (insertion sequence IS5376), MAGNESIUM ION, ... | | Authors: | Spinola-Amilibia, M, de la Gandara, A, Araujo-Bazan, L, Berger, J.M, Arias-Palomo, E. | | Deposit date: | 2022-09-20 | | Release date: | 2023-05-03 | | Method: | ELECTRON MICROSCOPY (3.35 Å) | | Cite: | IS21 family transposase cleaved donor complex traps two right-handed superhelical crossings.
Nat Commun, 14, 2023
|
|
6ZQU
 
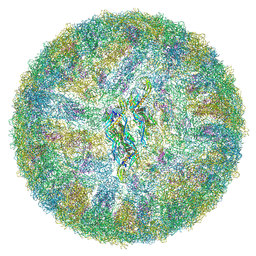 | | Cryo-EM structure of mature Dengue virus 2 at 3.1 angstrom resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Genome polyprotein | | Authors: | Renner, M, Dejnirattisai, W, Carrique, L, Serna Martin, I, Karia, D, Ilca, S.L, Ho, S.F, Kotecha, A, Keown, J.R, Mongkolsapaya, J, Screaton, G.R, Grimes, J.M. | | Deposit date: | 2020-07-10 | | Release date: | 2021-01-20 | | Last modified: | 2021-08-04 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Flavivirus maturation leads to the formation of an occupied lipid pocket in the surface glycoproteins.
Nat Commun, 12, 2021
|
|
8BVP
 
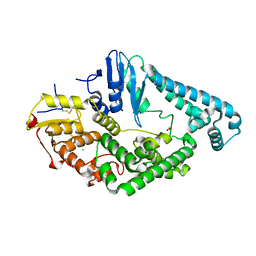 | | Crystal structure of an N-terminal fragment of the effector protein Lpg2504 (SidI) from Legionella pneumophila | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Restriction endonuclease | | Authors: | Machtens, D.A, Willerding, J.M, Eschenburg, S, Reubold, T.F. | | Deposit date: | 2022-12-05 | | Release date: | 2023-05-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of the N-terminal domain of the effector protein SidI of Legionella pneumophila reveals a glucosyl transferase domain.
Biochem.Biophys.Res.Commun., 661, 2023
|
|
8BWY
 
 | | In situ outer dynein arm from Chlamydomonas reinhardtii in a pre-power stroke state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Calmodulin, ... | | Authors: | Zimmermann, N.E.L, Noga, A, Obbineni, J.M, Ishikawa, T. | | Deposit date: | 2022-12-07 | | Release date: | 2023-05-10 | | Last modified: | 2023-06-28 | | Method: | ELECTRON MICROSCOPY (38 Å) | | Cite: | ATP-induced conformational change of axonemal outer dynein arms revealed by cryo-electron tomography.
Embo J., 42, 2023
|
|
8BX8
 
 | | In situ outer dynein arm from Chlamydomonas reinhardtii in the post-power stroke state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Dynein heavy chain, ... | | Authors: | Zimmermann, N.E.L, Noga, A, Obbineni, J.M, Ishikawa, T. | | Deposit date: | 2022-12-08 | | Release date: | 2023-05-10 | | Last modified: | 2023-06-28 | | Method: | ELECTRON MICROSCOPY (30.299999 Å) | | Cite: | ATP-induced conformational change of axonemal outer dynein arms revealed by cryo-electron tomography.
Embo J., 42, 2023
|
|
8BEI
 
 | |
8CGX
 
 | | structure of HEX-1 from N. crassa crystallized in cellulo, diffracted at 100K and resolved using XDS | | Descriptor: | Woronin body major protein | | Authors: | Boger, J, Schoenherr, R, Lahey-Rudolph, J.M, Harms, M, Kaiser, J, Nachtschatt, S, Wobbe, M, Koenig, P, Bourenkov, G, Schneider, T, Redecke, L. | | Deposit date: | 2023-02-06 | | Release date: | 2024-02-21 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | A streamlined approach to structure elucidation using in cellulo crystallized recombinant proteins, InCellCryst.
Nat Commun, 15, 2024
|
|
8CGY
 
 | | Trypanosoma brucei IMP dehydrogenase (ori) crystallized in High Five cells reveals native ligands ATP, GDP and phosphate. Diffraction data collection at 100 K in cellulo; XDS processing | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, GUANOSINE-5'-DIPHOSPHATE, Inosine-5'-monophosphate dehydrogenase, ... | | Authors: | Boger, J, Schoenherr, R, Lahey-Rudolph, J.M, Harms, M, Kaiser, J, Nachtschatt, S, Wobbe, M, Duden, R, Bourenkov, G, Schneider, T, Redecke, L. | | Deposit date: | 2023-02-06 | | Release date: | 2024-02-21 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | A streamlined approach to structure elucidation using in cellulo crystallized recombinant proteins, InCellCryst.
Nat Commun, 15, 2024
|
|
8CD5
 
 | | structure of HEX-1 from N. crassa crystallized in cellulo, diffracted at 100K and resolved using CrystFEL | | Descriptor: | Woronin body major protein | | Authors: | Boger, J, Schoenherr, R, Lahey-Rudolph, J.M, Harms, M, Kaiser, J, Nachtschatt, S, Wobbe, M, Koenig, P, Bourenkov, G, Schneider, T, Redecke, L. | | Deposit date: | 2023-01-30 | | Release date: | 2024-02-21 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | A streamlined approach to structure elucidation using in cellulo crystallized recombinant proteins, InCellCryst.
Nat Commun, 15, 2024
|
|
8C9J
 
 | | Crystal structure of human NQO1 by serial femtosecond crystallography | | Descriptor: | ACETATE ION, FLAVIN-ADENINE DINUCLEOTIDE, NAD(P)H dehydrogenase [quinone] 1 | | Authors: | Martin-Garcia, J.M, Grieco, A, Ruiz-Fresneda, M.A, Pacheco-Garcia, J.L, Pey, A, Botha, S, Ros, A. | | Deposit date: | 2023-01-23 | | Release date: | 2023-06-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Modular droplet injector for sample conservation providing new structural insight for the conformational heterogeneity in the disease-associated NQO1 enzyme.
Lab Chip, 23, 2023
|
|
2PHH
 
 | |
8C6D
 
 | | Production of antigenically stable enterovirus A71 virus-like particles in Pichia pastoris as a vaccine candidate. | | Descriptor: | (2S,3R,4E)-2-aminooctadec-4-ene-1,3-diol, Genome polyprotein, Genome polyprotein (Fragment) | | Authors: | Kingston, N.J, Snowden, J.S, Stonehouse, N.J, Rowlands, D.J, Hogle, J.M. | | Deposit date: | 2023-01-11 | | Release date: | 2023-02-22 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | Production of antigenically stable enterovirus A71 virus-like particles in Pichia pastoris as a vaccine candidate.
Biorxiv, 2023
|
|
1CTP
 
 | | STRUCTURE OF THE MAMMALIAN CATALYTIC SUBUNIT OF CAMP-DEPENDENT PROTEIN KINASE AND AN INHIBITOR PEPTIDE DISPLAYS AN OPEN CONFORMATION | | Descriptor: | MYRISTIC ACID, cAMP-DEPENDENT PROTEIN KINASE, cAMP-dependent protein kinase inhibitor, ... | | Authors: | Karlsson, R, Zheng, J, Xuong, N.H, Taylor, S.S, Sowadski, J.M. | | Deposit date: | 1993-04-08 | | Release date: | 1994-01-31 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure of the mammalian catalytic subunit of cAMP-dependent protein kinase and an inhibitor peptide displays an open conformation.
Acta Crystallogr.,Sect.D, 49, 1993
|
|
4XQD
 
 | | X-ray structure analysis of xylanase-WT at pH4.0 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Endo-1,4-beta-xylanase 2, IODIDE ION | | Authors: | Wan, Q, Park, J.M, Riccardi, D.M, Hanson, L.B, Fisher, Z, Smith, J.C, Ostermann, A, Schrader, T, Graham, D.E, Coates, L, Langan, P, Kovalevsky, A.Y. | | Deposit date: | 2015-01-19 | | Release date: | 2015-09-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Direct determination of protonation states and visualization of hydrogen bonding in a glycoside hydrolase with neutron crystallography.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
1CMK
 
 | | CRYSTAL STRUCTURES OF THE MYRISTYLATED CATALYTIC SUBUNIT OF CAMP-DEPENDENT PROTEIN KINASE REVEAL OPEN AND CLOSED CONFORMATIONS | | Descriptor: | IODIDE ION, MYRISTIC ACID, cAMP-DEPENDENT PROTEIN KINASE CATALYTIC SUBUNIT, ... | | Authors: | Zheng, J, Knighton, D.R, Xuong, N.-H, Taylor, S.S, Sowadski, J.M, Ten Eyck, L.F. | | Deposit date: | 1993-11-18 | | Release date: | 1994-05-31 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structures of the myristylated catalytic subunit of cAMP-dependent protein kinase reveal open and closed conformations.
Protein Sci., 2, 1993
|
|
4PXU
 
 | | Structural basis for the assembly of the mitotic motor kinesin-5 into bipolar tetramers | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Bipolar kinesin KRP-130 | | Authors: | Scholey, J.E, Nithianantham, S, Scholey, J.M, Al-Bassam, J. | | Deposit date: | 2014-03-25 | | Release date: | 2014-04-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.601 Å) | | Cite: | Structural basis for the assembly of the mitotic motor Kinesin-5 into bipolar tetramers.
Elife, 3, 2014
|
|
