8EUH
 
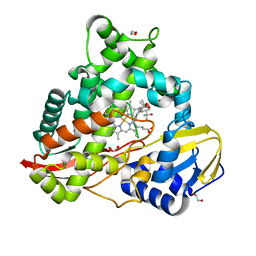 | | cytochrome P450terp (cyp108A1) bound to alpha-terpineol | | Descriptor: | 1,2-ETHANEDIOL, Cytochrome P450-terp, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Gable, J.A, Follmer, A.H, Poulos, T.L. | | Deposit date: | 2022-10-18 | | Release date: | 2023-02-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Cooperative Substrate Binding Controls Catalysis in Bacterial Cytochrome P450terp (CYP108A1).
J.Am.Chem.Soc., 2023
|
|
8EUK
 
 | |
8EUL
 
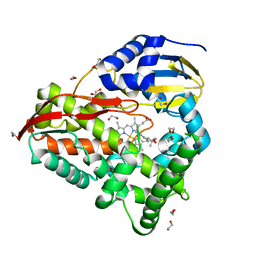 | | cytochrome P450terp (cyp108A1) mutant F188A bound to alpha-terpineol | | Descriptor: | 1,2-ETHANEDIOL, Cytochrome P450-terp, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Gable, J.A, Follmer, A.H, Poulos, T.L. | | Deposit date: | 2022-10-18 | | Release date: | 2023-02-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Cooperative Substrate Binding Controls Catalysis in Bacterial Cytochrome P450terp (CYP108A1).
J.Am.Chem.Soc., 2023
|
|
8ESW
 
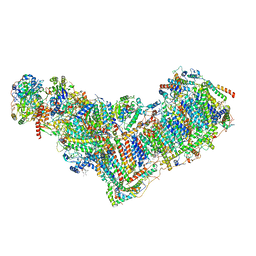 | | Structure of mitochondrial complex I from Drosophila melanogaster, Flexible-class 1 | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Padavannil, A, Letts, J.A. | | Deposit date: | 2022-10-15 | | Release date: | 2023-03-29 | | Last modified: | 2023-04-05 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Resting mitochondrial complex I from Drosophila melanogaster adopts a helix-locked state.
Elife, 12, 2023
|
|
8ESZ
 
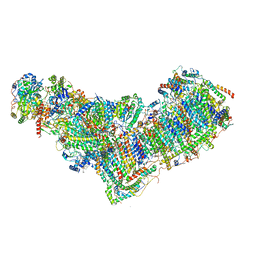 | | Structure of mitochondrial complex I from Drosophila melanogaster, Helix-locked state | | Descriptor: | (2R)-3-{[(S)-hydroxy(3-methylbutoxy)phosphoryl]oxy}-2-(octanoyloxy)propyl decanoate, 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, ... | | Authors: | Padavannil, A, Letts, J.A. | | Deposit date: | 2022-10-15 | | Release date: | 2023-03-29 | | Last modified: | 2023-04-05 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Resting mitochondrial complex I from Drosophila melanogaster adopts a helix-locked state.
Elife, 12, 2023
|
|
8EPQ
 
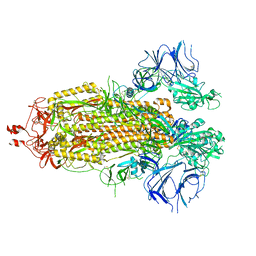 | |
8EPN
 
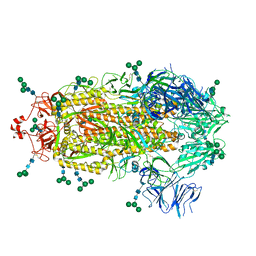 | |
8EPP
 
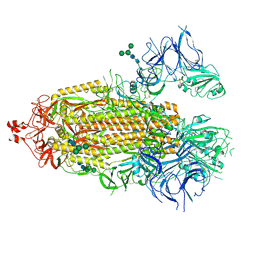 | |
3ESX
 
 | | E16KE61KD126KD150K Flavodoxin from Anabaena | | Descriptor: | CALCIUM ION, CHLORIDE ION, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Herguedas, B, Hermoso, J.A, Martinez-Julvez, M, Goni, G, Medina, M. | | Deposit date: | 2008-10-06 | | Release date: | 2009-02-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Flavodoxin: A compromise between efficiency and versatility in the electron transfer from Photosystem I to Ferredoxin-NADP(+) reductase
Biochim.Biophys.Acta, 1787, 2009
|
|
8ELU
 
 | | HRAS R97G Crystal form 1 | | Descriptor: | CALCIUM ION, GTPase HRas, MAGNESIUM ION, ... | | Authors: | Johnson, C.W, Rodrigues, J.A, Mattos, C. | | Deposit date: | 2022-09-26 | | Release date: | 2023-09-06 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Allosteric site variants affect GTP hydrolysis on Ras.
Protein Sci., 32, 2023
|
|
8EML
 
 | | Crystal Structure of Gsx2 Homeodomain in Complex with DNA | | Descriptor: | DNA (5'-D(P*GP*AP*GP*CP*TP*AP*AP*TP*TP*AP*AP*AP*GP*C)-3'), DNA (5'-D(P*GP*CP*TP*TP*TP*AP*AP*TP*TP*AP*GP*CP*TP*C)-3'), GS homeobox 2, ... | | Authors: | Webb, J.A, Kovall, R.A. | | Deposit date: | 2022-09-28 | | Release date: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Crystal Structure of Gsx2 Homeodomain in Complex with DNA
To Be Published
|
|
8FK3
 
 | |
8G0P
 
 | |
8G0Q
 
 | |
8G2E
 
 | | PKM2 bound to compound 2 | | Descriptor: | 1,2-ETHANEDIOL, 3-[(3-aminophenyl)methyl]-5-methyl-7-[methyl(oxidanyl)-$l^{3}-sulfanyl]pyridazino[4,5-b]indol-4-one, MAGNESIUM ION, ... | | Authors: | Stuckey, J.A. | | Deposit date: | 2023-02-03 | | Release date: | 2023-05-17 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.838 Å) | | Cite: | Development of Novel Small-Molecule Activators of Pyruvate Kinase Muscle Isozyme 2, PKM2, to Reduce Photoreceptor Apoptosis.
Pharmaceuticals, 16, 2023
|
|
8FAK
 
 | | DNA replication fork binding triggers structural changes in the PriA DNA helicase that regulate the PriA-PriB replication restart pathway in E. coli | | Descriptor: | DNA (5'-D(P*CP*AP*GP*AP*CP*TP*CP*AP*TP*TP*TP*AP*GP*CP*CP*CP*TP*TP*AP*TP*CP*CP*G)-3'), DNA (5'-D(P*CP*GP*GP*AP*TP*AP*AP*GP*GP*GP*CP*TP*GP*AP*GP*CP*AP*CP*GP*CP*CP*GP*A)-3'), DNA (5'-D(P*TP*CP*GP*GP*CP*GP*TP*GP*CP*TP*C)-3'), ... | | Authors: | Duckworth, A.T, Ducos, P.L, McMillan, S.D, Satyshur, K.A, Blumenthal, K.H, Deorio, H.R, Larson, J.A, Sandler, S.J, Grant, T, Keck, J.L. | | Deposit date: | 2022-11-28 | | Release date: | 2023-05-10 | | Last modified: | 2023-05-24 | | Method: | ELECTRON MICROSCOPY (3.22 Å) | | Cite: | Replication fork binding triggers structural changes in the PriA helicase that govern DNA replication restart in E. coli.
Nat Commun, 14, 2023
|
|
8G2V
 
 | | Cryo-EM structure of recombinant human LECT2 amyloid fibril core | | Descriptor: | Leukocyte cell-derived chemotaxin-2 | | Authors: | Richards, L.S, Flores, M.D, Zink, S, Schibrowsky, N.A, Sawaya, M.R, Rodriguez, J.A. | | Deposit date: | 2023-02-06 | | Release date: | 2023-08-16 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.715 Å) | | Cite: | Cryo-EM structure of a human LECT2 amyloid fibril reveals a network of polar ladders at its core.
Structure, 31, 2023
|
|
8FAC
 
 | |
8FEM
 
 | | Panicum vigratum Dihydroflavonol 4-reductase complexed with NADP | | Descriptor: | Dihydroflavonol 4-Reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Lewis, J.A, Kang, C. | | Deposit date: | 2022-12-06 | | Release date: | 2023-09-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Structural Similarities and Overlapping Activities among Dihydroflavonol 4-Reductase, Flavanone 4-Reductase, and Anthocyanidin Reductase Offer Metabolic Flexibility in the Flavonoid Pathway.
Int J Mol Sci, 24, 2023
|
|
8FEN
 
 | | Panicum vigratum Dihydroflavonol 4-reductase complexed with NADP and DHQ | | Descriptor: | (2R,3R)-2-(3,4-DIHYDROXYPHENYL)-3,5,7-TRIHYDROXY-2,3-DIHYDRO-4H-CHROMEN-4-ONE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Panicum virgatum Dihydroflavonol 4-Reductase | | Authors: | Lewis, J.A, Kang, C. | | Deposit date: | 2022-12-06 | | Release date: | 2023-09-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structural Similarities and Overlapping Activities among Dihydroflavonol 4-Reductase, Flavanone 4-Reductase, and Anthocyanidin Reductase Offer Metabolic Flexibility in the Flavonoid Pathway.
Int J Mol Sci, 24, 2023
|
|
8FFQ
 
 | | Wildtype rabbit TRPV5 into nanodiscs in the presence of PI(4,5)P2 and ruthenium red | | Descriptor: | 1-PALMITOYL-2-LINOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, ERGOSTEROL, Transient receptor potential cation channel subfamily V member 5, ... | | Authors: | De Jesus-Perez, J.J, Fluck, E.C, Pumroy, R.A, Protopopova, A.D, Rocereta, J.A, Moiseenkova-Bell, V.Y. | | Deposit date: | 2022-12-09 | | Release date: | 2024-02-07 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (2.65 Å) | | Cite: | Molecular details of ruthenium red pore block in TRPV channels.
Embo Rep., 25, 2024
|
|
8FFM
 
 | | Wildtype rat TRPV2 in nanodiscs bound to RR and 2-APB | | Descriptor: | 1,2-DIDECANOYL-SN-GLYCERO-3-PHOSPHOETHANOLAMINE, 2-aminoethyl diphenylborinate, Transient receptor potential cation channel subfamily V member 2, ... | | Authors: | Pumroy, R.A, Protopopova, A.D, Rocereta, J.A, De Jesus-Perez, J.J, Fluck, E.C, Moiseenkova-Bell, V.Y. | | Deposit date: | 2022-12-09 | | Release date: | 2024-02-07 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Molecular details of ruthenium red pore block in TRPV channels.
Embo Rep., 25, 2024
|
|
8FFL
 
 | | Wildtype rat TRPV2 in nanodiscs bound to RR | | Descriptor: | 1,2-DIDECANOYL-SN-GLYCERO-3-PHOSPHOETHANOLAMINE, Transient receptor potential cation channel subfamily V member 2 | | Authors: | Pumroy, R.A, Protopopova, A.D, Rocereta, J.A, De Jesus-Perez, J.J, Fluck, E.C, Moiseenkova-Bell, V.Y. | | Deposit date: | 2022-12-09 | | Release date: | 2024-02-07 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.47 Å) | | Cite: | Molecular details of ruthenium red pore block in TRPV channels.
Embo Rep., 25, 2024
|
|
8FFN
 
 | | RR-bound wildtype rabbit TRPV5 in nanodiscs | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, ERGOSTEROL, Transient receptor potential cation channel subfamily V member 5, ... | | Authors: | Fluck, E.C, De Jesus-Perez, J.J, Pumroy, R.A, Protopopova, A.D, Rocereta, J.A, Moiseenkova-Bell, V.Y. | | Deposit date: | 2022-12-09 | | Release date: | 2024-02-07 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (2.96 Å) | | Cite: | Molecular details of ruthenium red pore block in TRPV channels.
Embo Rep., 25, 2024
|
|
8G4J
 
 | |
