1O58
 
 | |
1O5U
 
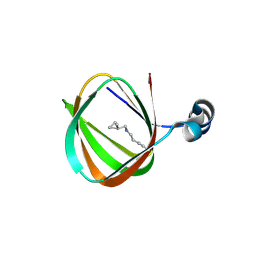 | |
3TOE
 
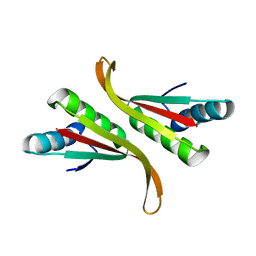 | | Structure of Mth10b | | Descriptor: | DNA/RNA-binding protein Alba | | Authors: | Pan, X.M, Zhang, N, Liu, Y.F, Liu, X. | | Deposit date: | 2011-09-05 | | Release date: | 2012-04-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.197 Å) | | Cite: | Molecular mechanism underlying the interaction of typical Sac10b family proteins with DNA.
Plos One, 7, 2012
|
|
6O6F
 
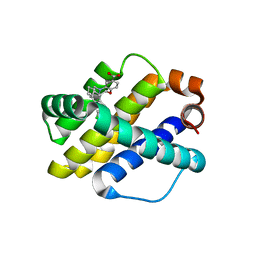 | | Co-crystal structure of Mcl1 with inhibitor | | Descriptor: | (3S)-5'-chloro-5-(cyclobutylmethyl)-2',3',4,5-tetrahydro-2H-spiro[1,5-benzoxazepine-3,1'-indene]-7-carboxylic acid, Induced myeloid leukemia cell differentiation protein Mcl-1 | | Authors: | Huang, X. | | Deposit date: | 2019-03-06 | | Release date: | 2019-05-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | AMG 176, a Selective MCL1 Inhibitor, Is Effective in Hematologic Cancer Models Alone and in Combination with Established Therapies.
Cancer Discov, 8, 2018
|
|
6OQD
 
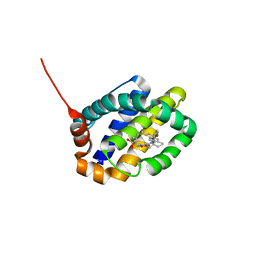 | | Crystal structure of Mcl1 with inhibitor 8 | | Descriptor: | (4S,7aR,9aR,10S,15R)-6'-chloro-10-hydroxy-15-methyl-3',4',7a,8,9,9a,10,11,12,13,14,15-dodecahydro-2'H,3H,5H-spiro[1,19-(ethanediylidene)-16lambda~6~-cyclobuta[i][1,4]oxazepino[3,4-f][1,2,7]thiadiazacyclohexadecine-4,1'-naphthalene]-16,16,18(7H,17H)-trione, Induced myeloid leukemia cell differentiation protein Mcl-1 | | Authors: | Huang, X. | | Deposit date: | 2019-04-26 | | Release date: | 2019-05-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | AMG 176, a Selective MCL1 Inhibitor, Is Effective in Hematologic Cancer Models Alone and in Combination with Established Therapies.
Cancer Discov, 8, 2018
|
|
6OQN
 
 | | Crystal structure of Mcl1 with inhibitor 7 | | Descriptor: | (4S)-5'-chloro-2',3',7,8,9,10,11,12-octahydro-3H,5H,14H-spiro[1,19-etheno-16lambda~6~-[1,4]oxazepino[3,4-i][1,4,5,10]oxathiadiazacyclohexadecine-4,1'-indene]-16,16,18(15H,17H)-trione, Induced myeloid leukemia cell differentiation protein Mcl-1 | | Authors: | Huang, X. | | Deposit date: | 2019-04-26 | | Release date: | 2019-05-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | AMG 176, a Selective MCL1 Inhibitor, Is Effective in Hematologic Cancer Models Alone and in Combination with Established Therapies.
Cancer Discov, 8, 2018
|
|
6OQB
 
 | | Co-crystal structure of Mcl1 with inhibitor 10 | | Descriptor: | (4S,7aR,9aR,10S,11E,15R)-6'-chloro-15-ethyl-10-hydroxy-3',4',7a,8,9,9a,10,13,14,15-decahydro-2'H,3H,5H-spiro[1,19-(ethanediylidene)-16lambda~6~-cyclobuta[i][1,4]oxazepino[3,4-f][1,2,7]thiadiazacyclohexadecine-4,1'-naphthalene]-16,16,18(7H,17H)-trione, Induced myeloid leukemia cell differentiation protein Mcl-1 | | Authors: | Huang, X. | | Deposit date: | 2019-04-26 | | Release date: | 2019-05-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | AMG 176, a Selective MCL1 Inhibitor, Is Effective in Hematologic Cancer Models Alone and in Combination with Established Therapies.
Cancer Discov, 8, 2018
|
|
6O6G
 
 | | Co-crystal structure of Mcl1 with inhibitor | | Descriptor: | (3S)-5-(cyclobutylmethyl)-3-(2,4-dichlorophenyl)-2,3,4,5-tetrahydro-1,5-benzoxazepine-7-carboxylic acid, Induced myeloid leukemia cell differentiation protein Mcl-1 | | Authors: | Huang, X. | | Deposit date: | 2019-03-06 | | Release date: | 2019-05-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | AMG 176, a Selective MCL1 Inhibitor, Is Effective in Hematologic Cancer Models Alone and in Combination with Established Therapies.
Cancer Discov, 8, 2018
|
|
7NA3
 
 | | HDM2 in complex with compound 62 | | Descriptor: | 3-[4-(5-chloropyridin-3-yl)-2-[(2S)-1-methoxypropan-2-yl]-3-{(1R)-1-[(1r,4R)-4-methylcyclohexyl]ethyl}-3H-imidazo[4,5-c]pyridin-6-yl]-1,2,4-oxadiazol-5(4H)-one, Isoform 11 of E3 ubiquitin-protein ligase Mdm2, SULFATE ION | | Authors: | Scapin, G. | | Deposit date: | 2021-06-19 | | Release date: | 2021-11-10 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Discovery of MK-4688 : an Efficient Inhibitor of the HDM2-p53 Protein-Protein Interaction.
J.Med.Chem., 64, 2021
|
|
7NA2
 
 | | HDM2 in complex with compound 56 | | Descriptor: | 3-[4-(5-chloropyridin-3-yl)-2-[(4aR,7aR)-hexahydrocyclopenta[b][1,4]oxazin-4(4aH)-yl]-3-{[(1r,4R)-4-methylcyclohexyl]methyl}-3H-imidazo[4,5-c]pyridin-6-yl]-1,2,4-oxadiazol-5(4H)-one, Isoform 11 of E3 ubiquitin-protein ligase Mdm2 | | Authors: | Scapin, G. | | Deposit date: | 2021-06-19 | | Release date: | 2021-11-10 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Discovery of MK-4688 : an Efficient Inhibitor of the HDM2-p53 Protein-Protein Interaction.
J.Med.Chem., 64, 2021
|
|
7NA1
 
 | | HDM2 in complex with compound 2 | | Descriptor: | 8-(1-benzothiophen-5-yl)-7-[(4-chlorophenyl)methyl]-6-{[(1R)-1-cyclopropylethyl]amino}-7H-purine-2-carboxylic acid, CITRIC ACID, E3 ubiquitin-protein ligase Mdm2, ... | | Authors: | Scapin, G. | | Deposit date: | 2021-06-19 | | Release date: | 2021-11-10 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Discovery of MK-4688 : an Efficient Inhibitor of the HDM2-p53 Protein-Protein Interaction.
J.Med.Chem., 64, 2021
|
|
7NA4
 
 | | HDM2 in complex with compound 63 | | Descriptor: | 3-[4-(5-chloropyridin-3-yl)-2-[(R)-cyclopropyl(ethoxy)methyl]-3-{(1R)-1-[(1r,4R)-4-methylcyclohexyl]ethyl}-3H-imidazo[4,5-c]pyridin-6-yl]-1,2,4-oxadiazol-5(4H)-one, CHLORIDE ION, GLYCEROL, ... | | Authors: | Scapin, G. | | Deposit date: | 2021-06-19 | | Release date: | 2021-11-10 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Discovery of MK-4688 : an Efficient Inhibitor of the HDM2-p53 Protein-Protein Interaction.
J.Med.Chem., 64, 2021
|
|
4XKB
 
 | | Crystal Structure of GENOMES UNCOUPLED 4 (GUN4) in Complex with Deuteroporphyrin IX | | Descriptor: | 3,3'-(3,7,12,17-tetramethylporphyrin-2,18-diyl)dipropanoic acid, Ycf53-like protein | | Authors: | Chen, X, Pu, H, Liu, L. | | Deposit date: | 2015-01-11 | | Release date: | 2015-05-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.501 Å) | | Cite: | Crystal Structures of GUN4 in Complex with Porphyrins.
Mol Plant, 8, 2015
|
|
4XKC
 
 | |
3JB4
 
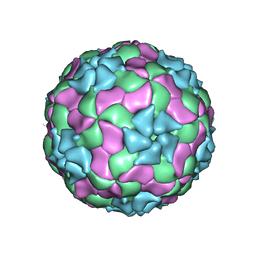 | | Structure of Ljungan virus: insight into picornavirus packaging | | Descriptor: | VP0, VP1, VP3 | | Authors: | Zhu, L, Wang, X.X, Ren, J.S, Porta, C, Wenham, H, Ekstrom, J.-O, Panjwani, A, Knowles, N.J, Kotecha, A, Siebert, A, Lindberg, M, Fry, E.E, Rao, Z.H, Tuthill, T.J, Stuart, D.I. | | Deposit date: | 2015-07-21 | | Release date: | 2015-10-21 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structure of Ljungan virus provides insight into genome packaging of this picornavirus.
Nat Commun, 6, 2015
|
|
1VJ2
 
 | |
1VJ1
 
 | |
1VKH
 
 | |
4L02
 
 | | Crystal Structure of SphK1 with inhibitor | | Descriptor: | (2R,4S)-1-[2-(4-{[4-(3,4-dichlorophenyl)-1,3-thiazol-2-yl]amino}phenyl)ethyl]-2-(hydroxymethyl)piperidin-4-ol, Sphingosine kinase 1 | | Authors: | Min, X, Walker, N, Wang, Z. | | Deposit date: | 2013-05-30 | | Release date: | 2013-07-24 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structure guided design of a series of sphingosine kinase (SphK) inhibitors.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
7YHS
 
 | | Structure of Csy-AcrIF4-dsDNA | | Descriptor: | AcrIF4, CRISPR type I-F/YPEST-associated protein Csy2, CRISPR-associated protein Csy3, ... | | Authors: | Feng, Y, Zhang, L.X. | | Deposit date: | 2022-07-14 | | Release date: | 2023-03-29 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.37 Å) | | Cite: | Anti-CRISPR protein AcrIF4 inhibits the type I-F CRISPR-Cas surveillance complex by blocking nuclease recruitment and DNA cleavage.
J.Biol.Chem., 298, 2022
|
|
6OVC
 
 | | hMcl1 inhibitor complex | | Descriptor: | (2S)-N-(benzylsulfonyl)-4-(cyclobutylmethyl)-2-(2,4-dichlorophenyl)-3,4-dihydro-2H-1,4-benzoxazine-6-carboxamide, Induced myeloid leukemia cell differentiation protein Mcl-1 | | Authors: | Poppe, L. | | Deposit date: | 2019-05-07 | | Release date: | 2019-05-22 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | AMG 176, a Selective MCL1 Inhibitor, Is Effective in Hematologic Cancer Models Alone and in Combination with Established Therapies.
Cancer Discov, 8, 2018
|
|
8W9B
 
 | | CryoEM structure of human PI3K-alpha (P85/P110-H1047R) with QR-8557 binding at an allosteric site | | Descriptor: | 1-[(1S)-1-(5-fluoranyl-3-methyl-1-benzofuran-2-yl)-2-methyl-propyl]-3-(1-oxidanylidene-2,3-dihydroisoindol-5-yl)urea, Phosphatidylinositol 3-kinase regulatory subunit alpha, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | Authors: | Huang, X, Ren, X, Zhong, W. | | Deposit date: | 2023-09-05 | | Release date: | 2024-04-17 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Cryo-EM structures reveal two allosteric inhibition modes of PI3K alpha H1047R involving a re-shaping of the activation loop.
Structure, 2024
|
|
8W9A
 
 | | CryoEM structure of human PI3K-alpha (P85/P110-H1047R) with QR-7909 binding at an allosteric site | | Descriptor: | 6-chloranyl-3-[[(1R)-1-[2-(1,3-dihydropyrrolo[3,4-c]pyridin-2-yl)-3,6-dimethyl-4-oxidanylidene-quinazolin-8-yl]ethyl]amino]pyridine-2-carboxylic acid, Phosphatidylinositol 3-kinase regulatory subunit alpha, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | Authors: | Huang, X, Ren, X, Zhong, W. | | Deposit date: | 2023-09-05 | | Release date: | 2024-04-17 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Cryo-EM structures reveal two allosteric inhibition modes of PI3K alpha H1047R involving a re-shaping of the activation loop.
Structure, 2024
|
|
6PU7
 
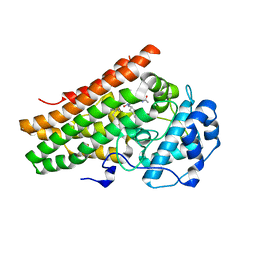 | | Human IDO1 in complex with compound 17 (N-{2-[(4-{N-[(7S)-4-fluorobicyclo[4.2.0]octa-1,3,5-trien-7-yl]-N'-hydroxycarbamimidoyl}-1,2,5-oxadiazol-3-yl)sulfanyl]ethyl}acetamide) | | Descriptor: | Indoleamine 2,3-dioxygenase 1, N-{2-[(4-{N-[(7S)-4-fluorobicyclo[4.2.0]octa-1,3,5-trien-7-yl]-N'-hydroxycarbamimidoyl}-1,2,5-oxadiazol-3-yl)sulfanyl]ethyl}acetamide, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Lesburg, C.A. | | Deposit date: | 2019-07-17 | | Release date: | 2019-12-04 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Discovery of Amino-cyclobutarene-derived Indoleamine-2,3-dioxygenase 1 (IDO1) Inhibitors for Cancer Immunotherapy.
Acs Med.Chem.Lett., 10, 2019
|
|
1Z92
 
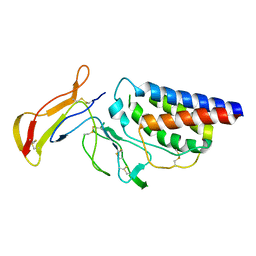 | | structure of interleukin-2 with its alpha receptor | | Descriptor: | Interleukin-2, Interleukin-2 receptor alpha chain | | Authors: | Rickert, M, Wang, X.Q, Goriatcheva, N, Boulanger, M.J, Garcia, K.C. | | Deposit date: | 2005-03-31 | | Release date: | 2005-06-07 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The structure of interleukin-2 complexed with its alpha receptor.
Science, 308, 2005
|
|
