3ZHI
 
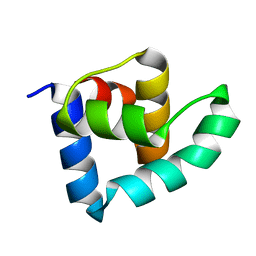 | | N-terminal domain of the CI repressor from bacteriophage TP901-1 | | Descriptor: | CI | | Authors: | Frandsen, K.H, Rasmussen, K.K, Poulsen, J.N, Lo Leggio, L. | | Deposit date: | 2012-12-21 | | Release date: | 2013-12-25 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Binding of the N-Terminal Domain of the Lactococcal Bacteriophage Tp901-1 Ci Repressor to its Target DNA: A Crystallography, Small Angle Scattering, and Nuclear Magnetic Resonance Study.
Biochemistry, 52, 2013
|
|
7CEL
 
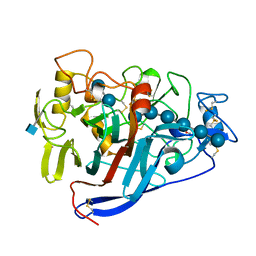 | | CBH1 (E217Q) IN COMPLEX WITH CELLOHEXAOSE AND CELLOBIOSE | | Descriptor: | 1,4-BETA-D-GLUCAN CELLOBIOHYDROLASE I, 2-acetamido-2-deoxy-beta-D-glucopyranose, COBALT (II) ION, ... | | Authors: | Divne, C, Stahlberg, J, Jones, T.A. | | Deposit date: | 1997-09-24 | | Release date: | 1997-12-24 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | High-resolution crystal structures reveal how a cellulose chain is bound in the 50 A long tunnel of cellobiohydrolase I from Trichoderma reesei.
J.Mol.Biol., 275, 1998
|
|
3LSA
 
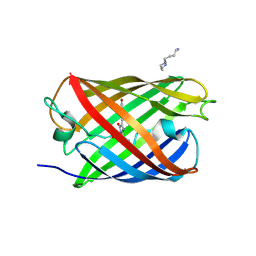 | | Padron0.9-OFF (non-fluorescent state) | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, GLYCEROL, Padron0.9, ... | | Authors: | Brakemann, T, Weber, G, Trowitzsch, S, Wahl, M.C, Jakobs, S. | | Deposit date: | 2010-02-12 | | Release date: | 2010-03-16 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Molecular basis of the light-driven switching of the photochromic fluorescent protein Padron.
J.Biol.Chem., 285, 2010
|
|
2G9V
 
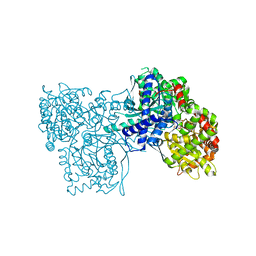 | | The crystal structure of glycogen phosphorylase in complex with (3R,4R,5R)-5-hydroxymethylpiperidine-3,4-diol and phosphate | | Descriptor: | 5-HYDROXYMETHYL-3,4-DIHYDROXYPIPERIDINE, Glycogen phosphorylase, muscle form, ... | | Authors: | Oikonomakos, N.G, Tiraidis, C, Leonidas, D.D, Zographos, S.E. | | Deposit date: | 2006-03-07 | | Release date: | 2007-01-16 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Iminosugars as potential inhibitors of glycogenolysis: structural insights into the molecular basis of glycogen phosphorylase inhibition.
J.Med.Chem., 49, 2006
|
|
1CLC
 
 | |
3LS3
 
 | | Padron0.9-ON (fluorescent state) | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, GLYCEROL, Padron0.9, ... | | Authors: | Brakemann, T, Weber, G, Trowitzsch, S, Wahl, M.C, Jakobs, S. | | Deposit date: | 2010-02-12 | | Release date: | 2010-03-16 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Molecular basis of the light-driven switching of the photochromic fluorescent protein Padron.
J.Biol.Chem., 285, 2010
|
|
1CB2
 
 | | CELLOBIOHYDROLASE II, CATALYTIC DOMAIN, MUTANT Y169F | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CELLOBIOHYDROLASE II, alpha-D-mannopyranose | | Authors: | Kleywegt, G.J, Szardenings, M, Jones, T.A. | | Deposit date: | 1995-11-25 | | Release date: | 1996-10-14 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The active site of Trichoderma reesei cellobiohydrolase II: the role of tyrosine 169.
Protein Eng., 9, 1996
|
|
7DSS
 
 | | Complex of FMDV and M8 Nab | | Descriptor: | M8 Nab, VP1 of O type FMDV capsid, VP2 of O-type FMDV capsid, ... | | Authors: | Dong, H, Liu, P. | | Deposit date: | 2021-01-02 | | Release date: | 2021-03-31 | | Last modified: | 2022-05-25 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural and molecular basis for foot-and-mouth disease virus neutralization by two potent protective antibodies.
Protein Cell, 13, 2022
|
|
7DST
 
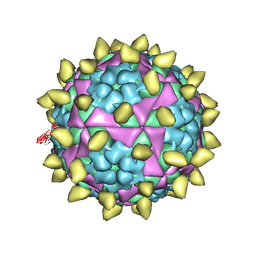 | | FMDV capsid in complex with M170 Nab | | Descriptor: | M170 Nab, VP1 of O type FMDV capsid, VP2 of O type FMDV capsid, ... | | Authors: | Dong, H, Liu, P. | | Deposit date: | 2021-01-02 | | Release date: | 2021-03-10 | | Last modified: | 2022-05-25 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural and molecular basis for foot-and-mouth disease virus neutralization by two potent protective antibodies.
Protein Cell, 13, 2022
|
|
2G9U
 
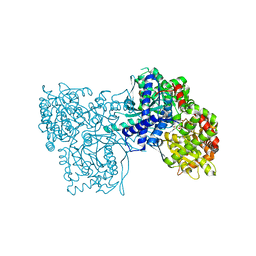 | | The crystal structure of glycogen phosphorylase in complex with (3R,4R,5R)-5-hydroxymethyl-1-(3-phenylpropyl)-piperidine-3,4-diol and phosphate | | Descriptor: | (3R,4R,5R)-5-(HYDROXYMETHYL)-1-(3-PHENYLPROPYL)PIPERIDINE-3,4-DIOL, Glycogen phosphorylase, muscle form, ... | | Authors: | Oikonomakos, N.G, Tiraidis, C, Leonidas, D.D, Zographos, S.E. | | Deposit date: | 2006-03-07 | | Release date: | 2007-01-16 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Iminosugars as potential inhibitors of glycogenolysis: structural insights into the molecular basis of glycogen phosphorylase inhibition.
J.Med.Chem., 49, 2006
|
|
2G9R
 
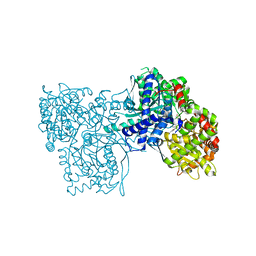 | | The crystal structure of glycogen phosphorylase b in complex with (3R,4R,5R)-5-hydroxymethyl-1-(3-phenylpropyl)-piperidine-3,4-diol | | Descriptor: | (3R,4R,5R)-5-(HYDROXYMETHYL)-1-(3-PHENYLPROPYL)PIPERIDINE-3,4-DIOL, Glycogen phosphorylase, muscle form | | Authors: | Oikonomakos, N.G, Tiraidis, C, Leonidas, D.D, Zographos, S.E. | | Deposit date: | 2006-03-07 | | Release date: | 2007-01-16 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Iminosugars as potential inhibitors of glycogenolysis: structural insights into the molecular basis of glycogen phosphorylase inhibition.
J.Med.Chem., 49, 2006
|
|
1HP8
 
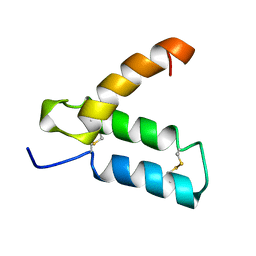 | | SOLUTION STRUCTURE OF HUMAN P8-MTCP1, A CYSTEINE-RICH PROTEIN ENCODED BY THE MTCP1 ONCOGENE,REVEALS A NEW ALPHA-HELICAL ASSEMBLY MOTIF, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | Cx9C motif-containing protein 4 | | Authors: | Barthe, P, Chiche, L, Strub, M.P, Roumestand, C. | | Deposit date: | 1997-08-26 | | Release date: | 1998-03-04 | | Last modified: | 2019-08-21 | | Method: | SOLUTION NMR | | Cite: | Solution structure of human p8MTCP1, a cysteine-rich protein encoded by the MTCP1 oncogene, reveals a new alpha-helical assembly motif.
J.Mol.Biol., 274, 1997
|
|
3ZDO
 
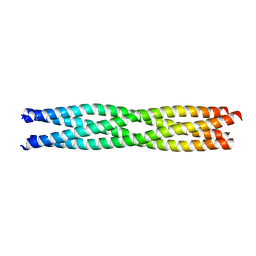 | | Tetramerization domain of Measles virus phosphoprotein | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, PHOSPHOPROTEIN | | Authors: | Communie, G, Crepin, T, Jensen, M.R, Blackledge, M, Ruigrok, R.W.H. | | Deposit date: | 2012-11-29 | | Release date: | 2013-04-24 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Structure of the Tetramerization Domain of Measles Virus Phosphoprotein.
J.Virol., 87, 2013
|
|
2YDL
 
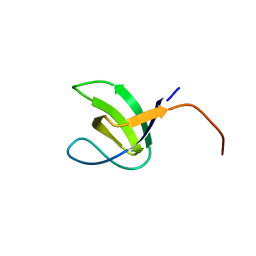 | | Crystal structure of SH3C from CIN85 | | Descriptor: | SH3 DOMAIN-CONTAINING KINASE-BINDING PROTEIN 1 | | Authors: | Bravo, J, Cardenes, N. | | Deposit date: | 2011-03-22 | | Release date: | 2012-03-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Distinct Ubiquitin Binding Modes Exhibited by SH3 Domains: Molecular Determinants and Functional Implications.
Plos One, 8, 2013
|
|
2BYW
 
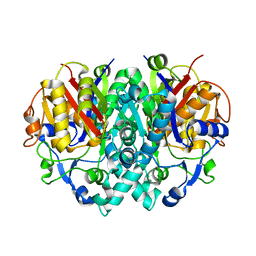 | |
2BYX
 
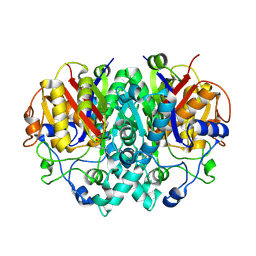 | |
1GZJ
 
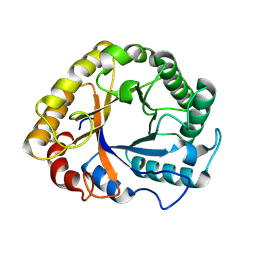 | |
3CEL
 
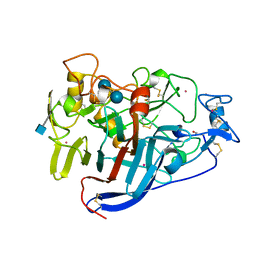 | | ACTIVE-SITE MUTANT E212Q DETERMINED AT PH 6.0 WITH CELLOBIOSE BOUND IN THE ACTIVE SITE | | Descriptor: | 1,4-BETA-D-GLUCAN CELLOBIOHYDROLASE I, 2-acetamido-2-deoxy-beta-D-glucopyranose, CADMIUM ION, ... | | Authors: | Divne, C, Stahlberg, J, Jones, T.A. | | Deposit date: | 1996-08-24 | | Release date: | 1997-03-12 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Activity studies and crystal structures of catalytically deficient mutants of cellobiohydrolase I from Trichoderma reesei.
J.Mol.Biol., 264, 1996
|
|
2Z2U
 
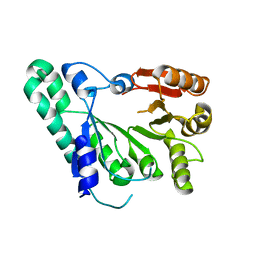 | | Crystal structure of archaeal TYW1 | | Descriptor: | UPF0026 protein MJ0257 | | Authors: | Suzuki, Y, Ishitani, R, Nureki, O. | | Deposit date: | 2007-05-28 | | Release date: | 2007-10-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of the Radical SAM Enzyme Catalyzing Tricyclic Modified Base Formation in tRNA
J.Mol.Biol., 372, 2007
|
|
2HU6
 
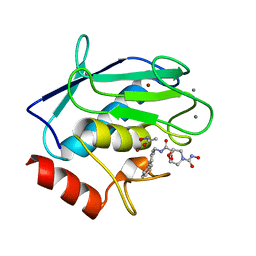 | | Crystal structure of human MMP-12 in complex with acetohydroxamic acid and a bicyclic inhibitor | | Descriptor: | (1S,5S,7R)-N~7~-(BIPHENYL-4-YLMETHYL)-N~3~-HYDROXY-6,8-DIOXA-3-AZABICYCLO[3.2.1]OCTANE-3,7-DICARBOXAMIDE, ACETOHYDROXAMIC ACID, CALCIUM ION, ... | | Authors: | Mannino, C, Nievo, M, Machetti, F, Papakyriakou, A, Calderone, V, Fragai, M, Guarna, A. | | Deposit date: | 2006-07-26 | | Release date: | 2006-12-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | Synthesis of bicyclic molecular scaffolds (BTAa): an investigation towards new selective MMP-12 inhibitors.
Bioorg.Med.Chem., 14, 2006
|
|
2E0M
 
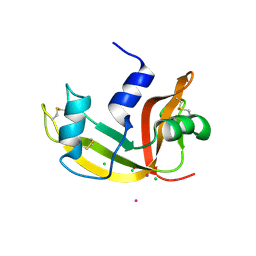 | | Mutant Human Ribonuclease 1 (T24L, Q28L, R31L, R32L) | | Descriptor: | CADMIUM ION, CHLORIDE ION, Ribonuclease | | Authors: | Yamada, H, Tamada, T, Kosaka, M, Kuroki, R. | | Deposit date: | 2006-10-10 | | Release date: | 2007-08-28 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | 'Crystal lattice engineering,' an approach to engineer protein crystal contacts by creating intermolecular symmetry: crystallization and structure determination of a mutant human RNase 1 with a hydrophobic interface of leucines
Protein Sci., 16, 2007
|
|
1E0V
 
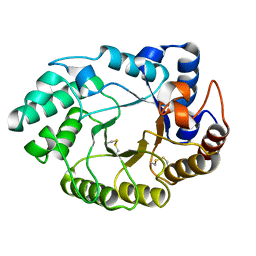 | | Xylanase 10A from Sreptomyces lividans. cellobiosyl-enzyme intermediate at 1.7 A | | Descriptor: | ENDO-1,4-BETA-XYLANASE A, beta-D-glucopyranose-(1-4)-2-deoxy-2-fluoro-alpha-D-glucopyranose | | Authors: | Ducros, V, Charnock, S.J, Derewenda, U, Derewenda, Z.S, Dauter, Z, Dupont, C, Shareck, F, Morosoli, R, Kluepfel, D, Davies, G.J. | | Deposit date: | 2000-04-10 | | Release date: | 2001-04-05 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Substrate Specificity in Glycoside Hydrolase Family 10. Structural and Kinetic Analysis of the Streptomyces Lividans Xylanase 10A
J.Biol.Chem., 275, 2000
|
|
1W9N
 
 | | Isolation and characterization of epilancin 15X, a novel antibiotic from a clinical strain of Staphylococcus epidermidis | | Descriptor: | EPILANCIN 15X | | Authors: | Ekkelenkamp, M, Hanssen, M.G.M, Hsu, S.-T.D, de Jong, A, Milatovic, D, Verhoef, J, van Nuland, N.A.J. | | Deposit date: | 2004-10-14 | | Release date: | 2005-04-01 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Isolation and structural characterization of epilancin 15X, a novel lantibiotic from a clinical strain of Staphylococcus epidermidis.
FEBS Lett., 579, 2005
|
|
3U2K
 
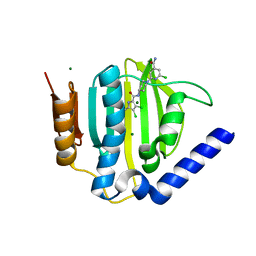 | | S. aureus GyrB ATPase domain in complex with a small molecule inhibitor | | Descriptor: | 2-chloro-6-(4-{[(3,4-dichloro-5-methyl-1H-pyrrol-2-yl)carbonyl]amino}piperidin-1-yl)pyridine-4-carboxamide, DNA gyrase subunit B, MAGNESIUM ION | | Authors: | Boriack-Sjodin, P.A, Prince, D.B, Eakin, A.E, Sherer, B.A. | | Deposit date: | 2011-10-03 | | Release date: | 2012-01-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Pyrrolamide DNA gyrase inhibitors: fragment-based nuclear magnetic resonance screening to identify antibacterial agents.
Antimicrob.Agents Chemother., 56, 2012
|
|
1E0W
 
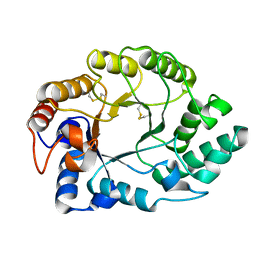 | | Xylanase 10A from Sreptomyces lividans. native structure at 1.2 angstrom resolution | | Descriptor: | ENDO-1,4-BETA-XYLANASE A | | Authors: | Ducros, V, Charnock, S.J, Derewenda, U, Derewenda, Z.S, Dauter, Z, Dupont, C, Shareck, F, Morosoli, R, Kluepfel, D, Davies, G.J. | | Deposit date: | 2000-04-10 | | Release date: | 2001-04-05 | | Last modified: | 2014-02-05 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Substrate Specificity in Glycoside Hydrolase Family 10. Structural and Kinetic Analysis of the Streptomyces Lividans Xylanase 10A
J.Biol.Chem., 275, 2000
|
|
