8D29
 
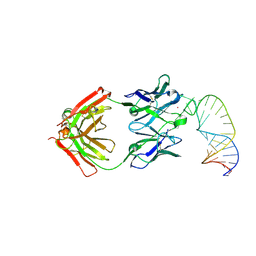 | |
1S1U
 
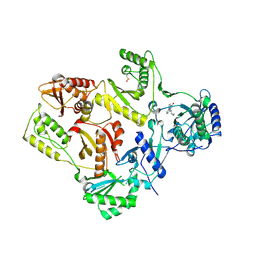 | | Crystal structure of L100I mutant HIV-1 reverse transcriptase in complex with nevirapine | | Descriptor: | 11-CYCLOPROPYL-5,11-DIHYDRO-4-METHYL-6H-DIPYRIDO[3,2-B:2',3'-E][1,4]DIAZEPIN-6-ONE, Reverse transcriptase | | Authors: | Ren, J, Nichols, C.E, Chamberlain, P.P, Stammers, D.K. | | Deposit date: | 2004-01-07 | | Release date: | 2004-06-29 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structures of HIV-1 reverse transcriptases mutated at codons 100, 106 and 108 and mechanisms of resistance to non-nucleoside inhibitors
J.Mol.Biol., 336, 2004
|
|
1S1W
 
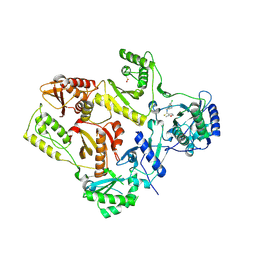 | | Crystal structure of V106A mutant HIV-1 reverse transcriptase in complex with UC-781 | | Descriptor: | 2-METHYL-FURAN-3-CARBOTHIOIC ACID [4-CHLORO-3-(3-METHYL-BUT-2-ENYLOXY)-PHENYL]-AMIDE, Reverse transcriptase | | Authors: | Ren, J, Nichols, C.E, Chamberlain, P.P, Stammers, D.K. | | Deposit date: | 2004-01-07 | | Release date: | 2004-06-29 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structures of HIV-1 reverse transcriptases mutated at codons 100, 106 and 108 and mechanisms of resistance to non-nucleoside inhibitors
J.Mol.Biol., 336, 2004
|
|
1S1T
 
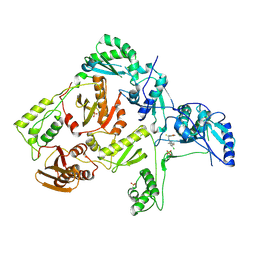 | | Crystal structure of L100I mutant HIV-1 reverse transcriptase in complex with UC-781 | | Descriptor: | 2-METHYL-FURAN-3-CARBOTHIOIC ACID [4-CHLORO-3-(3-METHYL-BUT-2-ENYLOXY)-PHENYL]-AMIDE, PHOSPHATE ION, Reverse transcriptase | | Authors: | Ren, J, Nichols, C.E, Chamberlain, P.P, Stammers, D.K. | | Deposit date: | 2004-01-07 | | Release date: | 2004-06-29 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structures of HIV-1 reverse transcriptases mutated at codons 100, 106 and 108 and mechanisms of resistance to non-nucleoside inhibitors
J.Mol.Biol., 336, 2004
|
|
8G8C
 
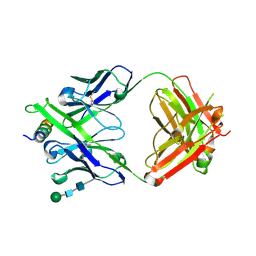 | | Crystal structure of DH1322.1 Fab in complex with HIV proximal MPER peptide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, DH1322.1 heavy chain, DH1322.1 light chain, ... | | Authors: | Niyongabo, A, Janus, B.M, Ofek, G. | | Deposit date: | 2023-02-17 | | Release date: | 2024-05-22 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Vaccine induction of heterologous HIV-1-neutralizing antibody B cell lineages in humans.
Cell, 187, 2024
|
|
8G8D
 
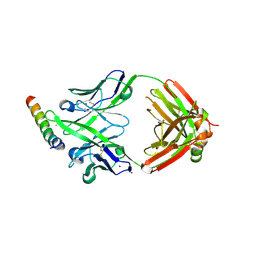 | | Crystal structure of DH1346 Fab in complex with HIV proximal MPER peptide | | Descriptor: | DH1346 heavy chain, DH1346 light chain, FLUORIDE ION, ... | | Authors: | Niyongabo, A, Janus, B.M, Ofek, G. | | Deposit date: | 2023-02-17 | | Release date: | 2024-05-22 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Vaccine induction of heterologous HIV-1-neutralizing antibody B cell lineages in humans.
Cell, 187, 2024
|
|
8G8A
 
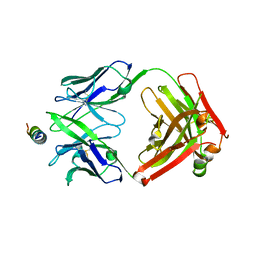 | | Crystal structure of DH1317.8 Fab in complex with HIV proximal MPER peptide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, DH1317.8 heavy chain, DH1317.8 light chain, ... | | Authors: | Janus, B.M, Astavans, A, Ofek, G. | | Deposit date: | 2023-02-17 | | Release date: | 2024-05-22 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Vaccine induction of heterologous HIV-1-neutralizing antibody B cell lineages in humans.
Cell, 187, 2024
|
|
1S1V
 
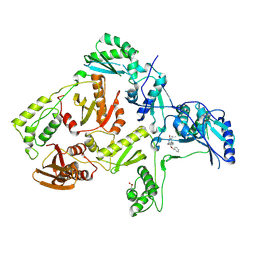 | | Crystal structure of L100I mutant HIV-1 reverse transcriptase in complex with TNK-651 | | Descriptor: | 6-BENZYL-1-BENZYLOXYMETHYL-5-ISOPROPYL URACIL, Reverse transcriptase | | Authors: | Ren, J, Nichols, C.E, Chamberlain, P.P, Stammers, D.K. | | Deposit date: | 2004-01-07 | | Release date: | 2004-06-29 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structures of HIV-1 reverse transcriptases mutated at codons 100, 106 and 108 and mechanisms of resistance to non-nucleoside inhibitors
J.Mol.Biol., 336, 2004
|
|
5KES
 
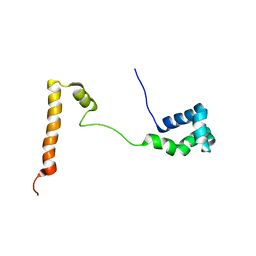 | | Solution structure of the yeast Ddi1 HDD domain | | Descriptor: | DNA damage-inducible protein 1 | | Authors: | Trempe, J.-F, Ratcliffe, C, Veverka, V, Saskova, K, Gehring, K. | | Deposit date: | 2016-06-10 | | Release date: | 2016-10-05 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural studies of the yeast DNA damage-inducible protein Ddi1 reveal domain architecture of this eukaryotic protein family.
Sci Rep, 6, 2016
|
|
1TKT
 
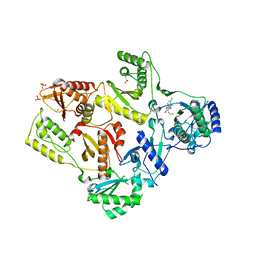 | | CRYSTAL STRUCTURE OF HIV-1 REVERSE TRANSCRIPTASE IN COMPLEX WITH GW426318 | | Descriptor: | 6-CHLORO-4-(CYCLOHEXYLOXY)-3-PROPYLQUINOLIN-2(1H)-ONE, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Hopkins, A.L, Ren, J, Stuart, D.I, Stammers, D.K. | | Deposit date: | 2004-06-09 | | Release date: | 2004-12-07 | | Last modified: | 2018-01-31 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Design of non-nucleoside inhibitors of HIV-1 reverse transcriptase with improved drug resistance properties. 1.
J.Med.Chem., 47, 2004
|
|
7QCW
 
 | |
5KL1
 
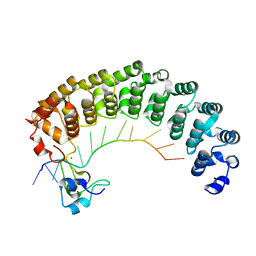 | | Crystal structure of the Pumilio-Nos-hunchback RNA complex | | Descriptor: | Maternal protein pumilio, Protein nanos, RNA (5'-R(*AP*AP*AP*UP*UP*GP*UP*AP*CP*AP*UP*A)-3'), ... | | Authors: | Qiu, C, Hall, T.M.T. | | Deposit date: | 2016-06-23 | | Release date: | 2016-08-17 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.701 Å) | | Cite: | Drosophila Nanos acts as a molecular clamp that modulates the RNA-binding and repression activities of Pumilio.
Elife, 5, 2016
|
|
1RT7
 
 | | HIV-1 REVERSE TRANSCRIPTASE COMPLEXED WITH UC84 | | Descriptor: | 1-METHYL ETHYL 1-CHLORO-5-[[(5,6DIHYDRO-2-METHYL-1,4-OXATHIIN-3-YL)CARBONYL]AMINO]BENZOATE, HIV-1 REVERSE TRANSCRIPTASE, PHOSPHATE ION | | Authors: | Ren, J, Stammers, D.K, Stuart, D.I. | | Deposit date: | 1998-07-29 | | Release date: | 1999-07-29 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structures of HIV-1 reverse transcriptase in complex with carboxanilide derivatives.
Biochemistry, 37, 1998
|
|
1RT6
 
 | | HIV-1 REVERSE TRANSCRIPTASE COMPLEXED WITH UC38 | | Descriptor: | 1-METHYL ETHYL 2-CHLORO-5-[[[(1-METHYLETHOXY)THIOOXO]METHYL]AMINO]-BENZOATE, HIV-1 REVERSE TRANSCRIPTASE, PHOSPHATE ION | | Authors: | Ren, J, Stammers, D.K, Stuart, D.I. | | Deposit date: | 1998-07-29 | | Release date: | 1999-07-29 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structures of HIV-1 reverse transcriptase in complex with carboxanilide derivatives.
Biochemistry, 37, 1998
|
|
1RT5
 
 | | HIV-1 REVERSE TRANSCRIPTASE COMPLEXED WITH UC10 | | Descriptor: | HIV-1 REVERSE TRANSCRIPTASE, N-[4-CLORO-3-(T-BUTYLOXOME)PHENYL-2-METHYL-3-FURAN-CARBOTHIAMIDE, PHOSPHATE ION | | Authors: | Ren, J, Stammers, D.K, Stuart, D.I. | | Deposit date: | 1998-07-29 | | Release date: | 1999-07-29 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structures of HIV-1 reverse transcriptase in complex with carboxanilide derivatives.
Biochemistry, 37, 1998
|
|
1RT4
 
 | | HIV-1 REVERSE TRANSCRIPTASE COMPLEXED WITH UC781 | | Descriptor: | 2-METHYL-FURAN-3-CARBOTHIOIC ACID [4-CHLORO-3-(3-METHYL-BUT-2-ENYLOXY)-PHENYL]-AMIDE, HIV-1 REVERSE TRANSCRIPTASE, PHOSPHATE ION | | Authors: | Ren, J, Stammers, D.K, Stuart, D.I. | | Deposit date: | 1998-07-29 | | Release date: | 1999-07-29 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structures of HIV-1 reverse transcriptase in complex with carboxanilide derivatives.
Biochemistry, 37, 1998
|
|
5KL8
 
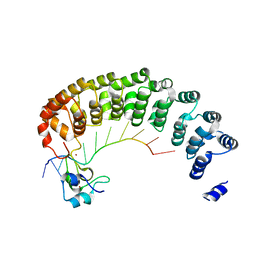 | | Crystal structure of the Pumilio-Nos-CyclinB RNA complex | | Descriptor: | Maternal protein pumilio, Protein nanos, RNA (5'-R(*UP*AP*UP*UP*UP*GP*UP*AP*AP*UP*U)-3'), ... | | Authors: | Qiu, C, Hall, T.M.T. | | Deposit date: | 2016-06-23 | | Release date: | 2016-08-17 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | Drosophila Nanos acts as a molecular clamp that modulates the RNA-binding and repression activities of Pumilio.
Elife, 5, 2016
|
|
1TL1
 
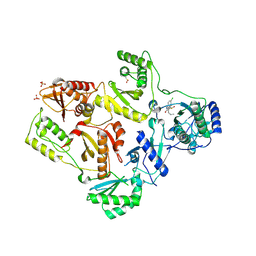 | | CRYSTAL STRUCTURE OF HIV-1 REVERSE TRANSCRIPTASE IN COMPLEX WITH GW451211 | | Descriptor: | 6-CHLORO-4-(CYCLOHEXYLSULFINYL)-3-PROPYLQUINOLIN-2(1H)-ONE, PHOSPHATE ION, Pol polyprotein, ... | | Authors: | Hopkins, A.L, Ren, J, Stuart, D.I, Stammers, D.K. | | Deposit date: | 2004-06-09 | | Release date: | 2004-12-07 | | Last modified: | 2022-12-21 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Design of non-nucleoside inhibitors of HIV-1 reverse transcriptase with improved drug resistance properties. 1.
J.Med.Chem., 47, 2004
|
|
5KLA
 
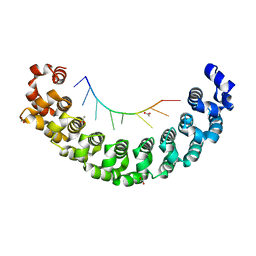 | |
1TL3
 
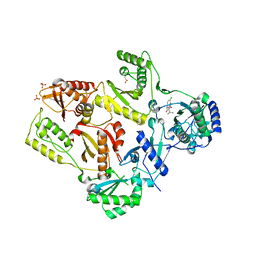 | | Crystal structure of hiv-1 reverse transcriptase in complex with gw450557 | | Descriptor: | 6-CHLORO-4-(CYCLOHEXYLOXY)-3-ISOPROPYLQUINOLIN-2(1H)-ONE, PHOSPHATE ION, Pol polyprotein, ... | | Authors: | Hopkins, A.L, Ren, J, Stuart, D.I, Stammers, D.K. | | Deposit date: | 2004-06-09 | | Release date: | 2004-12-07 | | Last modified: | 2022-12-21 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Design of non-nucleoside inhibitors of HIV-1 reverse transcriptase with improved drug resistance properties. 1.
J.Med.Chem., 47, 2004
|
|
1TKZ
 
 | | CRYSTAL STRUCTURE OF HIV-1 REVERSE TRANSCRIPTASE IN COMPLEX WITH GW429576 | | Descriptor: | 6-CHLORO-4-(CYCLOHEXYLSULFANYL)-3-PROPYLQUINOLIN-2(1H)-ONE, PHOSPHATE ION, Pol polyprotein, ... | | Authors: | Hopkins, A.L, Ren, J, Stuart, D.I, Stammers, D.K. | | Deposit date: | 2004-06-09 | | Release date: | 2004-12-07 | | Last modified: | 2020-01-15 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Design of non-nucleoside inhibitors of HIV-1 reverse transcriptase with improved drug resistance properties. 1.
J.Med.Chem., 47, 2004
|
|
1TKX
 
 | | CRYSTAL STRUCTURE OF HIV-1 REVERSE TRANSCRIPTASE IN COMPLEX WITH GW490745 | | Descriptor: | 4-[(CYCLOPROPYLETHYNYL)OXY]-6-FLUORO-3-ISOPROPYLQUINOLIN-2(1H)-ONE, Pol polyprotein, Reverse transcriptase, ... | | Authors: | Ren, J, Hopkins, A.L, Stuart, D.I, Stammers, D.K. | | Deposit date: | 2004-06-09 | | Release date: | 2004-12-07 | | Last modified: | 2022-12-21 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Design of non-nucleoside inhibitors of HIV-1 reverse transcriptase with improved drug resistance properties. 2.
J.Med.Chem., 47, 2004
|
|
6SOR
 
 | |
6SOO
 
 | |
6SOP
 
 | | Metal free structure of SynFtn variant E62A | | Descriptor: | CHLORIDE ION, Ferritin | | Authors: | Hemmings, A.M, Bradley, J.M. | | Deposit date: | 2019-08-29 | | Release date: | 2020-01-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Routes of iron entry into, and exit from, the catalytic ferroxidase sites of the prokaryotic ferritin SynFtn.
Dalton Trans, 49, 2020
|
|
