4RB0
 
 | | Crystal structure of Magnetospirillum gryphiswaldense MSR-1 SeMet-Apo-Fur | | Descriptor: | CITRATE ANION, DNA-binding transcriptional dual regulator of siderophore biosynthesis and transport(Fur family), SULFATE ION | | Authors: | Deng, Z, Chen, Z. | | Deposit date: | 2014-09-12 | | Release date: | 2015-07-15 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Mechanistic insights into metal ion activation and operator recognition by the ferric uptake regulator.
Nat Commun, 6
|
|
8R7R
 
 | |
6SLC
 
 | |
6LPI
 
 | | Crystal Structure of AHAS holo-enzyme | | Descriptor: | Acetolactate synthase isozyme 1 large subunit, Acetolactate synthase isozyme 1 small subunit, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Zhang, Y, Yang, X, Xi, Z, Shen, Y. | | Deposit date: | 2020-01-10 | | Release date: | 2020-11-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.849 Å) | | Cite: | Molecular architecture of the acetohydroxyacid synthase holoenzyme.
Biochem.J., 477, 2020
|
|
6RNV
 
 | |
4RB1
 
 | | Crystal structure of Magnetospirillum gryphiswaldense MSR-1 Fur-Mn2+-E. coli Fur box | | Descriptor: | DNA (5'-D(*CP*GP*CP*GP*AP*TP*AP*AP*TP*GP*AP*TP*AP*AP*TP*CP*AP*TP*TP*AP*TP*CP*CP*GP*C)-3'), DNA-binding transcriptional dual regulator of siderophore biosynthesis and transport(Fur family), MANGANESE (II) ION | | Authors: | Deng, Z, Chen, Z. | | Deposit date: | 2014-09-12 | | Release date: | 2015-07-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Mechanistic insights into metal ion activation and operator recognition by the ferric uptake regulator.
Nat Commun, 6
|
|
5CZA
 
 | | Yeast 20S proteasome beta5-D166N mutant | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, Probable proteasome subunit alpha type-7, ... | | Authors: | Huber, E.M, Groll, M. | | Deposit date: | 2015-07-31 | | Release date: | 2016-03-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A unified mechanism for proteolysis and autocatalytic activation in the 20S proteasome.
Nat Commun, 7, 2016
|
|
5CZ9
 
 | |
5CZ4
 
 | | Yeast 20S proteasome at 2.3 A resolution | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, Probable proteasome subunit alpha type-7, ... | | Authors: | Huber, E.M, Groll, M. | | Deposit date: | 2015-07-31 | | Release date: | 2016-03-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A unified mechanism for proteolysis and autocatalytic activation in the 20S proteasome.
Nat Commun, 7, 2016
|
|
5D0V
 
 | |
8PO3
 
 | |
8PO1
 
 | |
8PNZ
 
 | |
5D0S
 
 | |
5CZ6
 
 | | Yeast 20S proteasome beta5-T1A mutant in complex with Syringolin A, propeptide expressed in trans | | Descriptor: | (2S)-2-[[(2S)-1-[[(5S,8S,9E)-2,7-dioxo-5-propan-2-yl-1,6-diazacyclododeca-3,9-dien-8-yl]amino]-3-methyl-1-oxo-butan-2-yl]carbamoylamino]-3-methyl-butanoic acid, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Huber, E.M, Groll, M. | | Deposit date: | 2015-07-31 | | Release date: | 2016-03-23 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | A unified mechanism for proteolysis and autocatalytic activation in the 20S proteasome.
Nat Commun, 7, 2016
|
|
5DB0
 
 | | Menin in complex with MI-352 | | Descriptor: | 1-[(2R)-2,3-dihydroxypropyl]-5-[(4-{[6-(2,2,2-trifluoroethyl)thieno[2,3-d]pyrimidin-4-yl]amino}piperidin-1-yl)methyl]-1H-indole-2-carbonitrile, 1-[(2S)-2,3-dihydroxypropyl]-5-[(4-{[6-(2,2,2-trifluoroethyl)thieno[2,3-d]pyrimidin-4-yl]amino}piperidin-1-yl)methyl]-1 H-indole-2-carbonitrile, DIMETHYL SULFOXIDE, ... | | Authors: | Pollock, J, Dmitry, B, Cierpicki, T, Grembecka, J. | | Deposit date: | 2015-08-20 | | Release date: | 2016-03-30 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Property Focused Structure-Based Optimization of Small Molecule Inhibitors of the Protein-Protein Interaction between Menin and Mixed Lineage Leukemia (MLL).
J.Med.Chem., 59, 2016
|
|
5DB3
 
 | | Menin in complex with MI-574 | | Descriptor: | 6-methoxy-4-methyl-1-(1H-pyrazol-4-ylmethyl)-5-[(4-{[6-(2,2,2-trifluoroethyl)thieno[2,3-d]pyrimidin-4-yl]amino}piperidin-1-yl)methyl]-1H-indole-2-carbonitrile, DI(HYDROXYETHYL)ETHER, DIMETHYL SULFOXIDE, ... | | Authors: | Pollock, J, Borkin, D, Cierpicki, T, Grembecka, J. | | Deposit date: | 2015-08-20 | | Release date: | 2016-03-30 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Property Focused Structure-Based Optimization of Small Molecule Inhibitors of the Protein-Protein Interaction between Menin and Mixed Lineage Leukemia (MLL).
J.Med.Chem., 59, 2016
|
|
5DDG
 
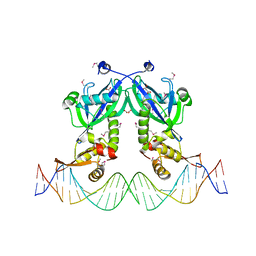 | | The structure of transcriptional factor AraR from Bacteroides thetaiotaomicron VPI in complex with target double strand DNA | | Descriptor: | DNA (27-MER), FORMIC ACID, MALONIC ACID, ... | | Authors: | Chang, C, Tesar, C, Rodionov, D, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2015-08-24 | | Release date: | 2015-09-09 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.06 Å) | | Cite: | A novel transcriptional regulator of L-arabinose utilization in human gut bacteria.
Nucleic Acids Res., 43, 2015
|
|
5DD4
 
 | | Apo structure of transcriptional factor AraR from Bacteroides thetaiotaomicron VPI | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, TRANSCRIPTIONAL REGULATOR AraR | | Authors: | Chang, C, Tesar, C, Rodionov, D, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2015-08-24 | | Release date: | 2015-09-09 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | A novel transcriptional regulator of L-arabinose utilization in human gut bacteria.
Nucleic Acids Res., 43, 2015
|
|
5D0T
 
 | | Yeast 20S proteasome beta5-D166N mutant in complex with MG132 | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, N-[(benzyloxy)carbonyl]-L-leucyl-N-[(2S)-1-hydroxy-4-methylpentan-2-yl]-L-leucinamide, ... | | Authors: | Huber, E.M, Groll, M. | | Deposit date: | 2015-08-03 | | Release date: | 2016-03-23 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A unified mechanism for proteolysis and autocatalytic activation in the 20S proteasome.
Nat Commun, 7, 2016
|
|
5CZ7
 
 | |
5D0X
 
 | | Yeast 20S proteasome beta5-T1S mutant in complex with Bortezomib | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, N-[(1R)-1-(DIHYDROXYBORYL)-3-METHYLBUTYL]-N-(PYRAZIN-2-YLCARBONYL)-L-PHENYLALANINAMIDE, ... | | Authors: | Huber, E.M, Groll, M. | | Deposit date: | 2015-08-03 | | Release date: | 2016-03-23 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A unified mechanism for proteolysis and autocatalytic activation in the 20S proteasome.
Nat Commun, 7, 2016
|
|
6C7R
 
 | | BRD4 BD1 in complex with compound CF53 | | Descriptor: | Bromodomain-containing protein 4, N-(3-cyclopropyl-1-methyl-1H-pyrazol-5-yl)-7-(3,5-dimethyl-1,2-oxazol-4-yl)-6-methoxy-2-methyl-9H-pyrimido[4,5-b]indol-4-amine | | Authors: | Meagher, J.L, Stuckey, J.A. | | Deposit date: | 2018-01-23 | | Release date: | 2018-08-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure-Based Discovery of CF53 as a Potent and Orally Bioavailable Bromodomain and Extra-Terminal (BET) Bromodomain Inhibitor.
J. Med. Chem., 61, 2018
|
|
8PO2
 
 | |
8PO0
 
 | |
