6YKZ
 
 | | Crystal structure of YTHDC1 with compound DHU_DC1_234 | | Descriptor: | SULFATE ION, YTHDC1, ~{N}-methyl-1,4,5,6-tetrahydrocyclopenta[c]pyrazole-3-carboxamide | | Authors: | Bedi, R.K, Huang, D, Wiedmer, L, Caflisch, A. | | Deposit date: | 2020-04-06 | | Release date: | 2020-07-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structure-based design of ligands of the m6A-RNA reader YTHDC1
Eur J Med Chem Rep, 5, 2022
|
|
6YL9
 
 | | Crystal structure of YTHDC1 with compound DHU_DC1_085 | | Descriptor: | 3-[(2~{R},5~{S})-2-(2,5-dimethylphenyl)-5-methyl-morpholin-4-yl]propane-1-sulfonamide, SULFATE ION, YTHDC1 | | Authors: | Bedi, R.K, Huang, D, Wiedmer, L, Caflisch, A. | | Deposit date: | 2020-04-06 | | Release date: | 2020-07-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure-based design of ligands of the m6A-RNA reader YTHDC1
Eur J Med Chem Rep, 5, 2022
|
|
6YNK
 
 | | Crystal structure of YTHDC1 with compound DHU_DC1_068 | | Descriptor: | 6-[[furan-2-ylmethyl(methyl)amino]methyl]-5~{H}-pyrimidine-2,4-dione, DI(HYDROXYETHYL)ETHER, SULFATE ION, ... | | Authors: | Bedi, R.K, Huang, D, Wiedmer, L, Caflisch, A. | | Deposit date: | 2020-04-13 | | Release date: | 2020-07-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structure-based design of ligands of the m6A-RNA reader YTHDC1
Eur J Med Chem Rep, 5, 2022
|
|
6YNN
 
 | | Crystal structure of YTHDC1 with compound DHU_DC1_135 | | Descriptor: | 6-[[(2-chloranyl-6-fluoranyl-phenyl)methyl-methyl-amino]methyl]-1~{H}-pyrimidine-2,4-dione, SULFATE ION, YTHDC1 | | Authors: | Bedi, R.K, Huang, D, Wiedmer, L, Caflisch, A. | | Deposit date: | 2020-04-14 | | Release date: | 2020-07-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Atomistic and Thermodynamic Analysis of N6-Methyladenosine (m 6 A) Recognition by the Reader Domain of YTHDC1.
J Chem Theory Comput, 17, 2021
|
|
6YOQ
 
 | |
6YNJ
 
 | | Crystal structure of YTHDC1 with compound DHU_DC1_046 | | Descriptor: | (2~{R})-2-(2-chlorophenyl)-5,5-dimethyl-morpholine, SULFATE ION, YTHDC1 | | Authors: | Bedi, R.K, Huang, D, Wiedmer, L, Caflisch, A. | | Deposit date: | 2020-04-13 | | Release date: | 2020-07-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure-based design of ligands of the m6A-RNA reader YTHDC1
Eur J Med Chem Rep, 5, 2022
|
|
6YNM
 
 | | Crystal structure of YTHDC1 with compound DHU_DC1_096 | | Descriptor: | 1-[(2~{R},5~{S})-2-(3-chlorophenyl)-5-methyl-morpholin-4-yl]-3-methoxy-propan-2-ol, SULFATE ION, YTHDC1 | | Authors: | Bedi, R.K, Huang, D, Wiedmer, L, Caflisch, A. | | Deposit date: | 2020-04-14 | | Release date: | 2020-07-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure-based design of ligands of the m6A-RNA reader YTHDC1
Eur J Med Chem Rep, 5, 2022
|
|
6YKJ
 
 | | Crystal structure of YTHDC1 with compound DHU_DC1_125 | | Descriptor: | SULFATE ION, YTHDC1, ~{N}-methyl-4-(methylamino)pyridine-2-carboxamide | | Authors: | Bedi, R.K, Huang, D, Wiedmer, L, Caflisch, A. | | Deposit date: | 2020-04-06 | | Release date: | 2020-07-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure-based design of ligands of the m6A-RNA reader YTHDC1
Eur J Med Chem Rep, 5, 2022
|
|
6YNI
 
 | | Crystal structure of YTHDC1 with compound DHU_DC1_036 | | Descriptor: | (2~{R},3~{R})-2-(3-chlorophenyl)-3,5,5-trimethyl-morpholine, SULFATE ION, YTHDC1 | | Authors: | Bedi, R.K, Huang, D, Wiedmer, L, Caflisch, A. | | Deposit date: | 2020-04-13 | | Release date: | 2020-07-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Structure-based design of ligands of the m6A-RNA reader YTHDC1
Eur J Med Chem Rep, 5, 2022
|
|
6YNL
 
 | | Crystal structure of YTHDC1 with compound DHU_DC1_078 | | Descriptor: | 3-[(2~{R})-5,5-dimethyl-2-phenyl-morpholin-4-yl]-~{N}-(methylcarbamoyl)propanamide, SULFATE ION, YTHDC1 | | Authors: | Bedi, R.K, Huang, D, Wiedmer, L, Caflisch, A. | | Deposit date: | 2020-04-14 | | Release date: | 2020-07-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure-based design of ligands of the m6A-RNA reader YTHDC1
Eur J Med Chem Rep, 5, 2022
|
|
6YNP
 
 | | Crystal structure of YTHDC1 with compound T96C1 | | Descriptor: | CHLORIDE ION, SULFATE ION, YTHDC1, ... | | Authors: | Bedi, R.K, Huang, D, Wiedmer, L, Caflisch, A. | | Deposit date: | 2020-04-14 | | Release date: | 2020-07-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Structure-based design of ligands of the m6A-RNA reader YTHDC1
Eur J Med Chem Rep, 5, 2022
|
|
6YL8
 
 | | Crystal structure of YTHDC1 with compound DHU_DC1_011 | | Descriptor: | (2~{R},5~{S})-2-(2-chlorophenyl)-5-methyl-morpholine, SULFATE ION, YTHDC1 | | Authors: | Bedi, R.K, Huang, D, Wiedmer, L, Caflisch, A. | | Deposit date: | 2020-04-06 | | Release date: | 2020-07-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure-based design of ligands of the m6A-RNA reader YTHDC1
Eur J Med Chem Rep, 5, 2022
|
|
6YNO
 
 | | Crystal structure of YTHDC1 with compound DHU_DC1_139 | | Descriptor: | 6-[[methyl-[(1-phenylpyrazol-3-yl)methyl]amino]methyl]-1~{H}-pyrimidine-2,4-dione, SULFATE ION, YTHDC1 | | Authors: | Bedi, R.K, Huang, D, Wiedmer, L, Caflisch, A. | | Deposit date: | 2020-04-14 | | Release date: | 2020-07-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Atomistic and Thermodynamic Analysis of N6-Methyladenosine (m 6 A) Recognition by the Reader Domain of YTHDC1.
J Chem Theory Comput, 17, 2021
|
|
6ZCN
 
 | | Crystal structure of YTHDC1 with m6A | | Descriptor: | N6-METHYLADENOSINE-5'-MONOPHOSPHATE, SULFATE ION, YTHDC1 | | Authors: | Bedi, R.K, Huang, D, Wiedmer, L, Caflisch, A. | | Deposit date: | 2020-06-11 | | Release date: | 2020-07-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Atomistic and Thermodynamic Analysis of N6-Methyladenosine (m 6 A) Recognition by the Reader Domain of YTHDC1.
J Chem Theory Comput, 17, 2021
|
|
6ZCM
 
 | | Crystal structure of YTHDC1 with compound DHU_DC1_180 | | Descriptor: | 6-[[cyclopropyl-[(7-methoxy-1,3-benzodioxol-5-yl)methyl]amino]methyl]-1~{H}-pyrimidine-2,4-dione, SULFATE ION, YTHDC1 | | Authors: | Bedi, R.K, Huang, D, Wiedmer, L, Caflisch, A. | | Deposit date: | 2020-06-11 | | Release date: | 2020-07-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | Atomistic and Thermodynamic Analysis of N6-Methyladenosine (m 6 A) Recognition by the Reader Domain of YTHDC1.
J Chem Theory Comput, 17, 2021
|
|
6ZOT
 
 | |
8SUX
 
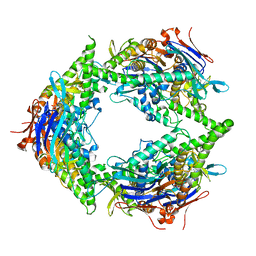 | | Structure of E. coli PtuA hexamer | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, PtuA | | Authors: | Shen, Z.F, Yang, X.Y, Fu, T.M. | | Deposit date: | 2023-05-13 | | Release date: | 2024-01-10 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.93 Å) | | Cite: | PtuA and PtuB assemble into an inflammasome-like oligomer for anti-phage defense.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8EEA
 
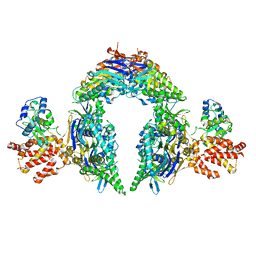 | | Structure of E.coli Septu (PtuAB) complex | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, PtuA, PtuB | | Authors: | Shen, Z.F, Fu, T.M. | | Deposit date: | 2022-09-06 | | Release date: | 2023-12-27 | | Last modified: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | PtuA and PtuB assemble into an inflammasome-like oligomer for anti-phage defense.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8EE7
 
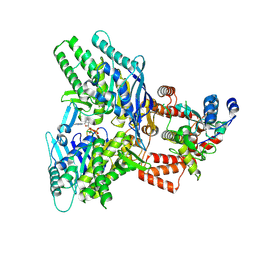 | |
8EE4
 
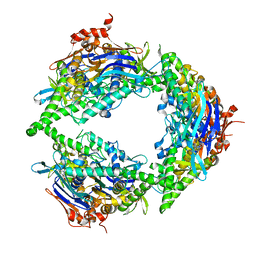 | | Structure of PtuA | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, PtuA | | Authors: | Shen, Z.F, Fu, T.M. | | Deposit date: | 2022-09-06 | | Release date: | 2024-01-03 | | Last modified: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | PtuA and PtuB assemble into an inflammasome-like oligomer for anti-phage defense.
Nat.Struct.Mol.Biol., 31, 2024
|
|
6ZD9
 
 | | Crystal structure of YTHDC1 apo purified using GST tag | | Descriptor: | DI(HYDROXYETHYL)ETHER, SULFATE ION, YTHDC1 | | Authors: | Bedi, R.K, Caflisch, A. | | Deposit date: | 2020-06-14 | | Release date: | 2020-07-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Atomistic and Thermodynamic Analysis of N6-Methyladenosine (m 6 A) Recognition by the Reader Domain of YTHDC1.
J Chem Theory Comput, 17, 2021
|
|
1TMJ
 
 | | Crystal structure of E.coli apo-HPPK(W89A) at 1.45 Angstrom resolution | | Descriptor: | 2-amino-4-hydroxy-6-hydroxymethyldihydropteridine pyrophosphokinase, CHLORIDE ION, MAGNESIUM ION | | Authors: | Blaszczyk, J, Ji, X. | | Deposit date: | 2004-06-10 | | Release date: | 2005-06-21 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Is the Critical Role of Loop 3 of Escherichia coli 6-Hydroxymethyl-7,8-dihydropterin Pyrophosphokinase in Catalysis Due to Loop-3 Residues Arginine-84 and Tryptophan-89? Site-Directed Mutagenesis, Biochemical, and Crystallographic Studies.
Biochemistry, 44, 2005
|
|
7VYQ
 
 | | Short chain dehydrogenase (SCR) cryoEM structure with NADP and ethyl 4-chloroacetoacetate | | Descriptor: | Carbonyl Reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ethyl 4-chloranyl-3-oxidanylidene-butanoate | | Authors: | Li, Y.H, Zhang, R.Z, Wang, C, Forouhar, F, Clarke, O, Vorobiev, S, Singh, S, Montelione, G.T, Szyperski, T, Xu, Y, Hunt, J.F. | | Deposit date: | 2021-11-15 | | Release date: | 2022-04-06 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.13 Å) | | Cite: | Oligomeric interactions maintain active-site structure in a noncooperative enzyme family.
Embo J., 41, 2022
|
|
6RT5
 
 | |
6RT6
 
 | |
