5KU3
 
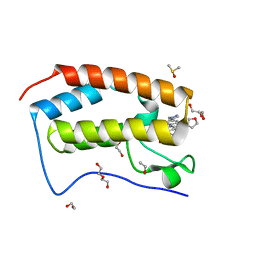 | | BRD4 bromodomain in complex with Cpd59 ((S)-1-(3-((2-fluoro-4-(1-methyl-1H-pyrazol-4-yl)phenyl)amino)-1-(tetrahydrofuran-3-yl)-6,7-dihydro-1H-pyrazolo[4,3-c]pyridin-5(4H)-yl)ethanone) | | Descriptor: | 1,2-ETHANEDIOL, 1-[3-[[2-fluoranyl-4-(1-methylpyrazol-4-yl)phenyl]amino]-1-[(3~{S})-oxolan-3-yl]-6,7-dihydro-4~{H}-pyrazolo[4,3-c]pyridin-5-yl]ethanone, Bromodomain-containing protein 4, ... | | Authors: | Murray, J.M, Huang, W. | | Deposit date: | 2016-07-12 | | Release date: | 2016-11-02 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.14 Å) | | Cite: | Discovery of a Potent and Selective in Vivo Probe (GNE-272) for the Bromodomains of CBP/EP300.
J. Med. Chem., 59, 2016
|
|
3BXV
 
 | |
2A9J
 
 | |
2RAC
 
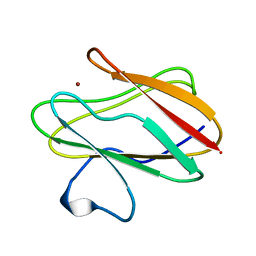 | | AMICYANIN REDUCED, PH 7.7, 1.3 ANGSTROMS | | Descriptor: | COPPER (I) ION, PROTEIN (AMICYANIN) | | Authors: | Cunane, L.M, Chen, Z.-W, Durley, R.C.E, Mathews, F.S. | | Deposit date: | 1998-10-02 | | Release date: | 1998-10-07 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Molecular basis for interprotein complex-dependent effects on the redox properties of amicyanin.
Biochemistry, 37, 1998
|
|
6ALC
 
 | | CREBBP bromodomain in complex with Cpd 4 (1-(1-(cyclopropylmethyl)-3-(1H-indol-4-yl)-1,4,6,7-tetrahydro-5H-pyrazolo[4,3-c]pyridin-5-yl)ethan-1-one) | | Descriptor: | 1,2-ETHANEDIOL, 1-[1-(cyclopropylmethyl)-3-(1H-indol-4-yl)-1,4,6,7-tetrahydro-5H-pyrazolo[4,3-c]pyridin-5-yl]ethan-1-one, CREB-binding protein, ... | | Authors: | Murray, J.M. | | Deposit date: | 2017-08-07 | | Release date: | 2018-08-08 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.391 Å) | | Cite: | Design and synthesis of a biaryl series as inhibitors for the bromodomains of CBP/P300.
Bioorg. Med. Chem. Lett., 28, 2018
|
|
1BHG
 
 | | HUMAN BETA-GLUCURONIDASE AT 2.6 A RESOLUTION | | Descriptor: | BETA-GLUCURONIDASE, alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-3)]alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Jain, S, Drendel, W.B. | | Deposit date: | 1996-03-04 | | Release date: | 1997-09-17 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Structure of human beta-glucuronidase reveals candidate lysosomal targeting and active-site motifs.
Nat.Struct.Biol., 3, 1996
|
|
4MBS
 
 | | Crystal Structure of the CCR5 Chemokine Receptor | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 4,4-difluoro-N-[(1S)-3-{(3-exo)-3-[3-methyl-5-(propan-2-yl)-4H-1,2,4-triazol-4-yl]-8-azabicyclo[3.2.1]oct-8-yl}-1-phenylpropyl]cyclohexanecarboxamide, Chimera protein of C-C chemokine receptor type 5 and Rubredoxin, ... | | Authors: | Tan, Q, Zhu, Y, Han, G.W, Li, J, Fenalti, G, Liu, H, Cherezov, V, Stevens, R.C, GPCR Network (GPCR), Zhao, Q, Wu, B. | | Deposit date: | 2013-08-19 | | Release date: | 2013-09-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Structure of the CCR5 chemokine receptor-HIV entry inhibitor maraviroc complex.
Science, 341, 2013
|
|
2RFX
 
 | | Crystal Structure of HLA-B*5701, presenting the self peptide, LSSPVTKSF | | Descriptor: | Beta-2-microglobulin, GLYCEROL, HLA class I histocompatibility antigen, ... | | Authors: | Archbold, J.K, Macdonald, W.A, Rossjohn, J, McCluskey, J. | | Deposit date: | 2007-10-02 | | Release date: | 2008-07-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Human leukocyte antigen class I-restricted activation of CD8+ T cells provides the immunogenetic basis of a systemic drug hypersensitivity
Immunity, 28, 2008
|
|
8PNM
 
 | | Structure of human KCTD15 BTB domain mutant G88D crystal form 2 | | Descriptor: | BTB/POZ domain-containing protein KCTD15 | | Authors: | Cruz Walma, D.A, Cros, J, Bradshaw, W, Richardson, W, Chen, Z, Chalk, R, Wilkie, A, Bullock, A.N. | | Deposit date: | 2023-06-30 | | Release date: | 2024-03-20 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | BTB domain mutations perturbing KCTD15 oligomerisation cause a distinctive frontonasal dysplasia syndrome.
J Med Genet, 61, 2024
|
|
8PNR
 
 | | Structure of human KCTD15 BTB domain mutant G88D crystal form 1 | | Descriptor: | BTB/POZ domain-containing protein KCTD15 | | Authors: | Cruz Walma, D.A, Cros, J, Bradshaw, W, Richardson, W, Chen, Z, Chalk, R, Wilkie, A, Bullock, A.N. | | Deposit date: | 2023-06-30 | | Release date: | 2024-03-20 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | BTB domain mutations perturbing KCTD15 oligomerisation cause a distinctive frontonasal dysplasia syndrome.
J Med Genet, 61, 2024
|
|
6JPJ
 
 | | Crystal structure of FGF401 in complex of FGFR4 | | Descriptor: | Fibroblast growth factor receptor 4, N-[5-cyano-4-(2-methoxyethylamino)pyridin-2-yl]-7-methanoyl-6-[(4-methyl-2-oxidanylidene-piperazin-1-yl)methyl]-3,4-dihydro-2H-1,8-naphthyridine-1-carboxamide, SULFATE ION | | Authors: | Zhou, Z, Chen, X, Chen, Y. | | Deposit date: | 2019-03-27 | | Release date: | 2019-05-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.638 Å) | | Cite: | Characterization of FGF401 as a reversible covalent inhibitor of fibroblast growth factor receptor 4.
Chem.Commun.(Camb.), 55, 2019
|
|
5J9U
 
 | | Crystal structure of the NuA4 core complex | | Descriptor: | Chromatin modification-related protein EAF6, Chromatin modification-related protein YNG2, Enhancer of polycomb-like protein 1, ... | | Authors: | Chen, Z.C, Xu, P. | | Deposit date: | 2016-04-11 | | Release date: | 2016-10-26 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | The NuA4 Core Complex Acetylates Nucleosomal Histone H4 through a Double Recognition Mechanism
Mol.Cell, 63, 2016
|
|
5IXS
 
 | | Lactate Dehydrogenase in complex with hydroxylactam inhibitor compound 9: (6R)-3-[(2-chlorophenyl)sulfanyl]-4-hydroxy-6-(3-hydroxyphenyl)-6-(thiophen-3-yl)-5,6-dihydropyridin-2(1H)-one | | Descriptor: | (6R)-3-[(2-chlorophenyl)sulfanyl]-4-hydroxy-6-(3-hydroxyphenyl)-6-(thiophen-3-yl)-5,6-dihydropyridin-2(1H)-one, 1,4,5,6-TETRAHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, ... | | Authors: | Ultsch, M, Eigenbrot, C. | | Deposit date: | 2016-03-23 | | Release date: | 2016-09-14 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Cell Active Hydroxylactam Inhibitors of Human Lactate Dehydrogenase with Oral Bioavailability in Mice.
Acs Med.Chem.Lett., 7, 2016
|
|
5J9T
 
 | | Crystal structure of the NuA4 core complex | | Descriptor: | Chromatin modification-related protein EAF6, Chromatin modification-related protein YNG2, Enhancer of polycomb-like protein 1, ... | | Authors: | Chen, Z.C, Xu, P. | | Deposit date: | 2016-04-11 | | Release date: | 2016-10-26 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The NuA4 Core Complex Acetylates Nucleosomal Histone H4 through a Double Recognition Mechanism
Mol.Cell, 63, 2016
|
|
5J9Q
 
 | | Crystal structure of the NuA4 core complex | | Descriptor: | Chromatin modification-related protein EAF6, Chromatin modification-related protein YNG2, Enhancer of polycomb-like protein 1, ... | | Authors: | Chen, Z.C, Xu, P. | | Deposit date: | 2016-04-11 | | Release date: | 2016-10-26 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | The NuA4 Core Complex Acetylates Nucleosomal Histone H4 through a Double Recognition Mechanism
Mol.Cell, 63, 2016
|
|
5BXJ
 
 | | Complex of the Fk1 domain mutant A19T of FKBP51 with 4-Nitrophenol | | Descriptor: | P-NITROPHENOL, Peptidyl-prolyl cis-trans isomerase FKBP5 | | Authors: | Wu, D, Tao, X, Chen, Z, Han, J, Jia, W, Li, X, Wang, Z, He, Y.X. | | Deposit date: | 2015-06-09 | | Release date: | 2016-05-25 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | The environmental endocrine disruptor p-nitrophenol interacts with FKBP51, a positive regulator of androgen receptor and inhibits androgen receptor signaling in human cells
J. Hazard. Mater., 307, 2016
|
|
5HC1
 
 | |
5J9W
 
 | | Crystal structure of the NuA4 core complex | | Descriptor: | ACETYL COENZYME *A, Chromatin modification-related protein EAF6, Chromatin modification-related protein YNG2, ... | | Authors: | Chen, Z.C, Xu, P. | | Deposit date: | 2016-04-11 | | Release date: | 2016-10-26 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The NuA4 Core Complex Acetylates Nucleosomal Histone H4 through a Double Recognition Mechanism
Mol.Cell, 63, 2016
|
|
5HBZ
 
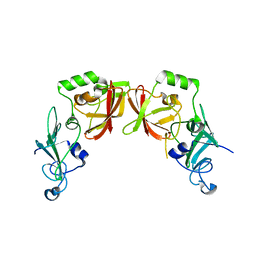 | |
6M5U
 
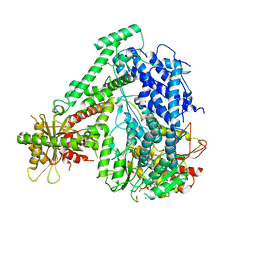 | | The coordinates of the monomeric terminase complex in the presence of the ADP-BeF3 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, MAGNESIUM ION, ... | | Authors: | Yang, Y.X, Yang, P, Wang, N, Zhu, L, Zhou, Z.H, Rao, Z.H, Wang, X.X. | | Deposit date: | 2020-03-11 | | Release date: | 2020-10-28 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Architecture of the herpesvirus genome-packaging complex and implications for DNA translocation.
Protein Cell, 11, 2020
|
|
6IEC
 
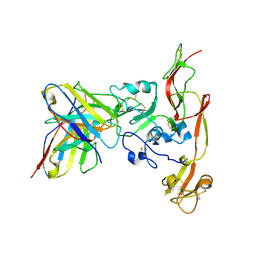 | | Structure of RVFV Gn and human monoclonal antibody R17 | | Descriptor: | NSmGnGc, R17 H chain, R17 L chain | | Authors: | Wang, Q.H, Wu, Y, Gao, F, Qi, J.X, Gao, G.F. | | Deposit date: | 2018-09-13 | | Release date: | 2019-04-10 | | Last modified: | 2019-07-10 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Neutralization mechanism of human monoclonal antibodies against Rift Valley fever virus.
Nat Microbiol, 4, 2019
|
|
5XFJ
 
 | | Crystal structure of LY2874455 in complex of FGFR4 gatekeeper mutation (V550M) | | Descriptor: | 2-[4-[E-2-[5-[(1R)-1-[3,5-bis(chloranyl)pyridin-4-yl]ethoxy]-1H-indazol-3-yl]ethenyl]pyrazol-1-yl]ethanol, Fibroblast growth factor receptor 4 | | Authors: | Wu, D, Chen, Y. | | Deposit date: | 2017-04-10 | | Release date: | 2018-08-08 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | LY2874455 potently inhibits FGFR gatekeeper mutants and overcomes mutation-based resistance.
Chem. Commun. (Camb.), 54, 2018
|
|
3VFV
 
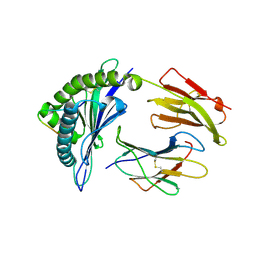 | | crystal structure of HLA B*3508 LPEP-P9Ala, peptide mutant P9-ala | | Descriptor: | Beta-2-microglobulin, LPEP peptide from EBV, P9A, ... | | Authors: | Liu, Y.C, Rossjohn, J, Gras, S. | | Deposit date: | 2012-01-10 | | Release date: | 2012-02-22 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | The Energetic Basis Underpinning T-cell Receptor Recognition of a Super-bulged Peptide Bound to a Major Histocompatibility Complex Class I Molecule.
J.Biol.Chem., 287, 2012
|
|
3VFW
 
 | | crystal structure of HLA B*3508 LPEP-P10Ala, peptide mutant P10-ala | | Descriptor: | Beta-2-microglobulin, LPEP peptide from EBV, P10A, ... | | Authors: | Liu, Y.C, Rossjohn, J, Gras, S. | | Deposit date: | 2012-01-10 | | Release date: | 2012-02-22 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The Energetic Basis Underpinning T-cell Receptor Recognition of a Super-bulged Peptide Bound to a Major Histocompatibility Complex Class I Molecule.
J.Biol.Chem., 287, 2012
|
|
3VFT
 
 | | crystal structure of HLA B*3508LPEP-P6Ala, peptide mutant P6-ala | | Descriptor: | Beta-2-microglobulin, LPEP peptide from EBV, P6A, ... | | Authors: | Liu, Y.C, Rossjohn, J, Gras, S. | | Deposit date: | 2012-01-10 | | Release date: | 2012-02-22 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.947 Å) | | Cite: | The Energetic Basis Underpinning T-cell Receptor Recognition of a Super-bulged Peptide Bound to a Major Histocompatibility Complex Class I Molecule.
J.Biol.Chem., 287, 2012
|
|
