3H2Y
 
 | | Crystal structure of YqeH GTPase from Bacillus anthracis with dGDP bound | | Descriptor: | 2'-DEOXYGUANOSINE-5'-DIPHOSPHATE, GTPase family protein | | Authors: | Brunzelle, J.S, Anderson, S.M, Xu, X, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-04-15 | | Release date: | 2009-06-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of YqeH GTPase from Bacillus anthracis with dGDP Bound
To be Published
|
|
3FF1
 
 | | Structure of Glucose 6-phosphate Isomerase from Staphylococcus aureus | | Descriptor: | GLUCOSE-6-PHOSPHATE, Glucose-6-phosphate isomerase, SODIUM ION | | Authors: | Anderson, S.M, Brunzelle, J.S, Onopriyenko, O, Peterson, S, Anderson, W.F, Savchenko, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2008-12-01 | | Release date: | 2009-02-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structure of Glucose 6-phosphate Isomerase from Staphylococcus aureus
TO BE PUBLISHED
|
|
3DR6
 
 | | Structure of yncA, a putative ACETYLTRANSFERASE from Salmonella typhimurium | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, yncA | | Authors: | Singer, A.U, Skarina, T, Onopriyenko, O, Edwards, A.M, Anderson, W.F, Savchenko, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2008-07-10 | | Release date: | 2008-09-09 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Funded by the national institute of
allergy and infectious diseases of nih (contract number
hhsn272200700058c).
To be Published
|
|
3HFR
 
 | | Crystal structure of glutamate racemase from Listeria monocytogenes | | Descriptor: | CHLORIDE ION, Glutamate racemase, O-(O-(2-AMINOPROPYL)-O'-(2-METHOXYETHYL)POLYPROPYLENE GLYCOL 500) | | Authors: | Majorek, K.A, Chruszcz, M, Zimmerman, M.D, Klimecka, M.M, Cymborowski, M, Skarina, T, Onopriyenko, O, Stam, J, Otwinowski, Z, Anderson, W.F, Savchenko, A, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-05-12 | | Release date: | 2009-06-09 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of glutamate racemase from Listeria monocytogenes
TO BE PUBLISHED
|
|
3HJV
 
 | | 1.7 Angstrom resolution crystal structure of an acyl carrier protein S-malonyltransferase from Vibrio cholerae O1 biovar eltor str. N16961 | | Descriptor: | 3-CYCLOHEXYL-1-PROPYLSULFONIC ACID, CHLORIDE ION, Malonyl Coa-acyl carrier protein transacylase, ... | | Authors: | Halavaty, A.S, Wawrzak, Z, Anderson, S, Skarina, T, Onopriyenko, O, Kwon, K, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-05-22 | | Release date: | 2009-06-09 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | 1.7 Angstrom resolution crystal structure of an acyl carrier protein S-malonyltransferase from Vibrio cholerae O1 biovar eltor str. N16961
To be Published
|
|
3R8Y
 
 | | Structure of the Bacillus anthracis tetrahydropicolinate succinyltransferase | | Descriptor: | 2,3,4,5-tetrahydropyridine-2,6-dicarboxylate N-acetyltransferase, CALCIUM ION | | Authors: | Anderson, S.M, Wawrzak, Z, Onopriyenko, O, Peterson, S.N, Anderson, W.F, Savchenko, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-03-24 | | Release date: | 2011-04-20 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of the Bacillus anthracis tetrahydropicolinate succinyltransferase
TO BE PUBLISHED
|
|
3T7Y
 
 | | Structure of an autocleavage-inactive mutant of the cytoplasmic domain of CT091, the YscU homologue of Chlamydia trachomatis | | Descriptor: | CHLORIDE ION, FORMIC ACID, SODIUM ION, ... | | Authors: | Singer, A.U, Wawrzak, Z, Skarina, T, Saikali, P, Anderson, W.F, Savchenko, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-07-31 | | Release date: | 2011-11-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of an autocleavage-inactive mutant of the cytoplasmic domain of CT091, the YscU homologue of Chlamydia trachomatis
TO BE PUBLISHED
|
|
3T7B
 
 | | Crystal Structure of N-acetyl-L-glutamate kinase from Yersinia pestis | | Descriptor: | Acetylglutamate kinase, GLUTAMIC ACID, S,R MESO-TARTARIC ACID | | Authors: | Demas, M.W, Solberg, R.G, Cooper, D.R, Chruszcz, M, Porebski, P.J, Zheng, H, Onopriyenko, O, Skarina, T, Savchenko, A, Anderson, W.F, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-07-29 | | Release date: | 2011-09-14 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of N-acetyl-L-glutamate kinase from Yersinia pestis
To be Published
|
|
3TL2
 
 | | Crystal structure of Bacillus anthracis str. Ames malate dehydrogenase in closed conformation. | | Descriptor: | 1,2-ETHANEDIOL, Malate dehydrogenase, THIOCYANATE ION | | Authors: | Blus, B.J, Chruszcz, M, Tkaczuk, K.L, Osinski, T, Cymborowski, M, Kudritska, M, Grimshaw, S, Savchenko, A, Anderson, W.F, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-08-29 | | Release date: | 2011-09-21 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of Bacillus anthracis str. Ames malate dehydrogenase in closed conformation.
TO BE PUBLISHED
|
|
3R3S
 
 | | Structure of the YghA Oxidoreductase from Salmonella enterica | | Descriptor: | FORMIC ACID, MAGNESIUM ION, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Anderson, S.M, Wawrzak, Z, Onopriyenko, O, Peterson, S.N, Anderson, W.F, Savchenko, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-03-16 | | Release date: | 2011-03-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Structure of the YghA Oxidoreductase from Salmonella enterica
TO BE PUBLISHED
|
|
3QYF
 
 | | Crystal structure of the CRISPR-associated protein SSO1393 from Sulfolobus solfataricus | | Descriptor: | CRISPR-ASSOCIATED PROTEIN, MAGNESIUM ION | | Authors: | Petit, P, Xu, X, Chang, C, Savchenko, A, Yakunin, A.F. | | Deposit date: | 2011-03-03 | | Release date: | 2011-03-23 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the CRISPR-associated protein SSO1393 from Sulfolobus solfataricus
To be Published
|
|
3QYJ
 
 | |
3QY1
 
 | | 1.54A Resolution Crystal Structure of a Beta-Carbonic Anhydrase from Salmonella enterica subsp. enterica serovar Typhimurium str. LT2 | | Descriptor: | Carbonic anhydrase, ZINC ION | | Authors: | Brunzelle, J.S, Wawrzak, Z, Onopriyenko, O, Anderson, W.F, Savchenko, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-03-02 | | Release date: | 2011-03-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | 1.54A Resolution Crystal Structure of a Beta-Carbonic Anhydrase from Salmonella enterica subsp. enterica serovar Typhimurium str. LT2
To be Published
|
|
3RUY
 
 | | Crystal Structure of the Ornithine-oxo acid transaminase RocD from Bacillus anthracis | | Descriptor: | Ornithine aminotransferase | | Authors: | Anderson, S.M, Wawrzak, Z, Brunzelle, J.S, Onopriyenko, O, Peterson, S.N, Anderson, W.F, Savchenko, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-05-05 | | Release date: | 2011-05-18 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Crystal Structure of the Ornithine-oxo acid transaminase RocD from Bacillus anthracis
To be Published
|
|
3R3R
 
 | | Structure of the YrdA ferripyochelin binding protein from Salmonella enterica | | Descriptor: | ZINC ION, ferripyochelin binding protein | | Authors: | Anderson, S.M, Wawrzak, Z, Onopriyenko, O, Peterson, S.N, Anderson, W.F, Savchenko, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-03-16 | | Release date: | 2011-03-30 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structure of the YrdA ferripyochelin binding protein from Salmonella enterica
TO BE PUBLISHED
|
|
3T5P
 
 | | Crystal structure of a putative diacylglycerol kinase from Bacillus anthracis str. Sterne | | Descriptor: | BmrU protein, MAGNESIUM ION | | Authors: | Hou, J, Zheng, H, Chruszcz, M, Cooper, D.R, Onopriyenko, O, Grimshaw, S, Savchenko, A, Anderson, W.F, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-07-27 | | Release date: | 2011-09-07 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of a putative diacylglycerol kinase from Bacillus anthracis str. Sterne
To be Published
|
|
3R0P
 
 | | Crystal structure of L-PSP putative endoribonuclease from uncultured organism | | Descriptor: | L-PSP putative endoribonuclease | | Authors: | Cuff, M.E, Petit, P, Xu, X, Cui, H, Savchenko, A, Yakunin, A.F. | | Deposit date: | 2011-03-08 | | Release date: | 2011-03-23 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of L-PSP putative endoribonuclease from uncultured organism
To be Published
|
|
3TYS
 
 | | Crystal structure of transcriptional regulator VanUg, Form II | | Descriptor: | Predicted transcriptional regulator | | Authors: | Stogios, P.J, Evdokimova, E, Wawrzak, Z, Depardieu, F, Courvalin, P, Shabalin, I, Chruszcz, M, Minor, W, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-09-26 | | Release date: | 2011-10-12 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.121 Å) | | Cite: | Crystal structure of transcriptional regulator VanUg, Form II
TO BE PUBLISHED
|
|
3T32
 
 | | Crystal structure of a putative C-S lyase from Bacillus anthracis | | Descriptor: | Aminotransferase, class I/II | | Authors: | Anderson, S.M, Wawrzak, Z, Gordon, E, Peterson, S.N, Porebski, P, Minor, W, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-07-24 | | Release date: | 2011-08-10 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of a putative C-S lyase from Bacillus anthracis
TO BE PUBLISHED
|
|
3SS6
 
 | | Crystal structure of the Bacillus anthracis acetyl-CoA acetyltransferase | | Descriptor: | Acetyl-CoA acetyltransferase, POTASSIUM ION, SULFATE ION | | Authors: | Anderson, S.M, Wawrzak, Z, Onopriyenko, O, Peterson, S.N, Anderson, W.F, Savchenko, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-07-07 | | Release date: | 2011-07-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of the Bacillus anthracis acetyl-CoA acetyltransferase
TO BE PUBLISHED
|
|
3TAU
 
 | | Crystal Structure of a Putative Guanylate Monophosphaste Kinase from Listeria monocytogenes EGD-e | | Descriptor: | Guanylate kinase, SODIUM ION, SULFATE ION | | Authors: | Brunzelle, J.S, Wawrzak, Z, Onopriyenko, O, Kwok, J, Anderson, W.F, Savchenko, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-08-04 | | Release date: | 2011-08-24 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal Structure of a Putative Guanylate Monophosphaste Kinase from Listeria monocytogenes EGD-e
To be Published
|
|
3TYK
 
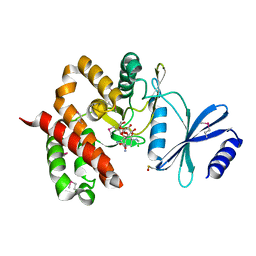 | | Crystal structure of aminoglycoside phosphotransferase APH(4)-Ia | | Descriptor: | CHLORIDE ION, HYGROMYCIN B VARIANT, Hygromycin-B 4-O-kinase | | Authors: | Stogios, P.J, Shabalin, I.G, Shakya, T, Evdokmova, E, Fan, Y, Chruszcz, M, Minor, W, Wright, G.D, Savchenko, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-09-26 | | Release date: | 2011-10-12 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure and function of APH(4)-Ia, a hygromycin B resistance enzyme.
J.Biol.Chem., 286, 2011
|
|
3UM9
 
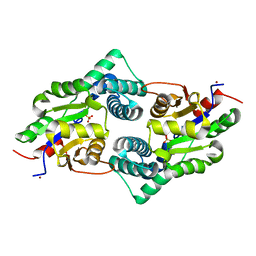 | | Crystal Structure of the Defluorinating L-2-Haloacid Dehalogenase Bpro0530 | | Descriptor: | Haloacid dehalogenase, type II, NICKEL (II) ION, ... | | Authors: | Chan, P.W.Y, Savchenko, A, Yakunin, A.F, Edwards, E.A, Pai, E.F. | | Deposit date: | 2011-11-12 | | Release date: | 2012-11-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Structural adaptations of L-2-haloacid dehalogenases that enable hydrolytic defluorination
To be Published
|
|
3UDO
 
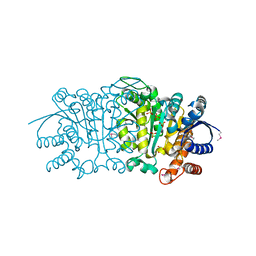 | | Crystal structure of putative isopropylamlate dehydrogenase from Campylobacter jejuni | | Descriptor: | 1,2-ETHANEDIOL, 3-isopropylmalate dehydrogenase, SULFATE ION | | Authors: | Tkaczuk, K.L, Chruszcz, M, Blus, B.J, Onopriyenko, O, Grimshaw, S, Savchenko, A, Anderson, W.F, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-10-28 | | Release date: | 2011-11-09 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of putative isopropylamlate dehydrogenase from Campylobacter jejuni
To be Published
|
|
3UUW
 
 | | 1.63 Angstrom Resolution Crystal Structure of Dehydrogenase (MviM) from Clostridium difficile. | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Minasov, G, Wawrzak, Z, Kudritska, M, Grimshaw, S, Papazisi, L, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-11-28 | | Release date: | 2011-12-21 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | 1.63 Angstrom Resolution Crystal Structure of Dehydrogenase (MviM) from Clostridium difficile.
TO BE PUBLISHED
|
|
