5YXA
 
 | | Crystal structure of the C-terminal fragment of NS1 protein from yellow fever virus | | Descriptor: | Non-structural protein 1 | | Authors: | Wang, H, Song, H, Qi, J, Shi, Y, Gao, G.F. | | Deposit date: | 2017-12-04 | | Release date: | 2018-01-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of the C-terminal fragment of NS1 protein from yellow fever virus.
Sci China Life Sci, 60, 2017
|
|
5XXQ
 
 | |
5XJC
 
 | | Cryo-EM structure of the human spliceosome just prior to exon ligation at 3.6 angstrom | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Zhang, X, Yan, C, Hang, J, Finci, I.L, Lei, J, Shi, Y. | | Deposit date: | 2017-04-30 | | Release date: | 2017-07-05 | | Last modified: | 2020-10-14 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | An Atomic Structure of the Human Spliceosome
Cell, 169, 2017
|
|
5Z58
 
 | | Cryo-EM structure of a human activated spliceosome (early Bact) at 4.9 angstrom. | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, BUD13 homolog, Cell division cycle 5-like protein, ... | | Authors: | Zhang, X, Yan, C, Zhan, X, Li, L, Lei, J, Shi, Y. | | Deposit date: | 2018-01-17 | | Release date: | 2018-09-19 | | Last modified: | 2020-10-14 | | Method: | ELECTRON MICROSCOPY (4.9 Å) | | Cite: | Structure of the human activated spliceosome in three conformational states.
Cell Res., 28, 2018
|
|
5Z57
 
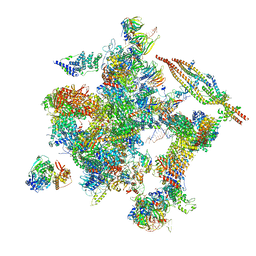 | | Cryo-EM structure of the human activated spliceosome (late Bact) at 6.5 angstrom | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, ALANINE, BUD13 homolog, ... | | Authors: | Zhang, X, Yan, C, Zhan, X, Li, L, Lei, J, Shi, Y. | | Deposit date: | 2018-01-17 | | Release date: | 2018-09-19 | | Last modified: | 2020-10-14 | | Method: | ELECTRON MICROSCOPY (6.5 Å) | | Cite: | Structure of the human activated spliceosome in three conformational states.
Cell Res., 28, 2018
|
|
1IY6
 
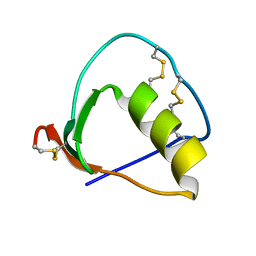 | | Solution structure of OMSVP3 variant, P14C/N39C | | Descriptor: | OMSVP3 | | Authors: | Hemmi, H, Kumazaki, T, Yamazaki, T, Kojima, S, Yoshida, T, Kyogoku, Y, Katsu, M, Yokosawa, H, Miura, K, Kobayashi, Y. | | Deposit date: | 2002-07-23 | | Release date: | 2003-03-11 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Inhibitory Specificity Change of Ovomucoid Third Domain of the Silver Pheasant upon Introduction of an Engineered Cys14-Cys39 Bond
BIOCHEMISTRY, 42, 2003
|
|
1IY5
 
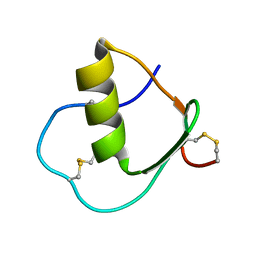 | | Solution structure of wild type OMSVP3 | | Descriptor: | OMSVP3 | | Authors: | Hemmi, H, Kumazaki, T, Yamazaki, T, Kojima, S, Yoshida, T, Kyogoku, Y, Katsu, M, Yokosawa, H, Miura, K, Kobayashi, Y. | | Deposit date: | 2002-07-23 | | Release date: | 2003-03-11 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Inhibitory Specificity Change of Ovomucoid Third Domain of the Silver Pheasant upon Introduction of an Engineered Cys14-Cys39 Bond
BIOCHEMISTRY, 42, 2003
|
|
1DVV
 
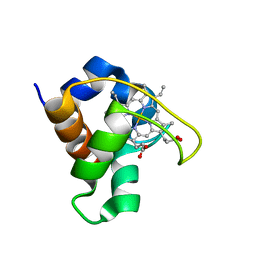 | | SOLUTION STRUCTURE OF THE QUINTUPLE MUTANT OF CYTOCHROME C-551 FROM PSEUDOMONAS AERUGINOSA | | Descriptor: | CYTOCHROME C551, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Hasegawa, J, Uchiyama, S, Tanimoto, Y, Mizutani, M, Kobayashi, Y, Sambongi, Y, Igarashi, Y. | | Deposit date: | 2000-01-22 | | Release date: | 2000-11-29 | | Last modified: | 2024-04-10 | | Method: | SOLUTION NMR | | Cite: | Selected mutations in a mesophilic cytochrome c confer the stability of a thermophilic counterpart.
J.Biol.Chem., 275, 2000
|
|
7EXS
 
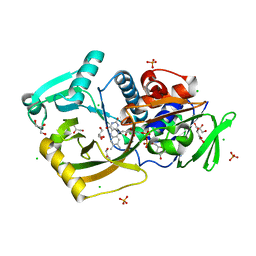 | | Thermomicrobium roseum sarcosine oxidase mutant - S320R | | Descriptor: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Xin, Y, Shen, C, Tang, M.W, Shi, Y, Guo, Z.T, Gu, Z.H, Shao, J, Zhang, L. | | Deposit date: | 2021-05-28 | | Release date: | 2021-07-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Recreating the natural evolutionary trend in key microdomains provides an effective strategy for engineering of a thermomicrobial N-demethylase.
J.Biol.Chem., 298, 2022
|
|
1IYQ
 
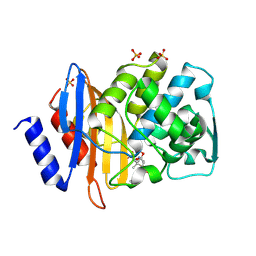 | | Toho-1 beta-Lactamase In Complex With Benzylpenicillin | | Descriptor: | OPEN FORM - PENICILLIN G, SULFATE ION, Toho-1 beta-lactamase | | Authors: | Shimamura, T, Ibuka, A, Fushinobu, S, Wakagi, T, Ishiguro, M, Ishii, Y, Matsuzawa, H. | | Deposit date: | 2002-09-04 | | Release date: | 2002-12-11 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Acyl-intermediate Structures of the Extended-spectrum Class A beta -Lactamase, Toho-1, in Complex with Cefotaxime, Cephalothin, and Benzylpenicillin.
J.Biol.Chem., 277, 2002
|
|
5HWS
 
 | |
1J2E
 
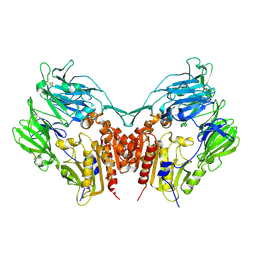 | | Crystal structure of Human Dipeptidyl peptidase IV | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Dipeptidyl peptidase IV | | Authors: | Hiramatsu, H, Kyono, K, Higashiyama, Y, Fukushima, C, Shima, H, Sugiyama, S, Inaka, K, Yamamoto, A, Shimizu, R. | | Deposit date: | 2002-12-30 | | Release date: | 2003-12-30 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The structure and function of human dipeptidyl peptidase IV, possessing a unique eight-bladed beta-propeller fold.
Biochem.Biophys.Res.Commun., 302, 2003
|
|
1IYP
 
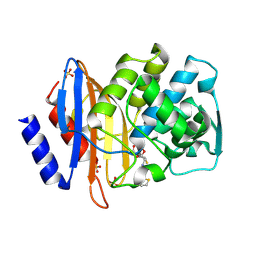 | | Toho-1 beta-Lactamase In Complex With Cephalothin | | Descriptor: | CEPHALOTHIN GROUP, SULFATE ION, Toho-1 beta-lactamase | | Authors: | Shimamura, T, Ibuka, A, Fushinobu, S, Wakagi, T, Ishiguro, M, Ishii, Y, Matsuzawa, H. | | Deposit date: | 2002-09-04 | | Release date: | 2002-12-11 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Acyl-intermediate Structures of the Extended-spectrum Class A beta -Lactamase, Toho-1, in Complex with Cefotaxime, Cephalothin, and Benzylpenicillin.
J.Biol.Chem., 277, 2002
|
|
7EVN
 
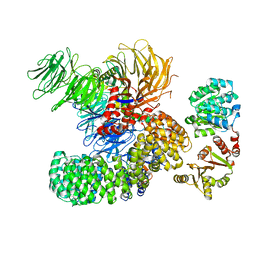 | | The cryo-EM structure of the DDX42-SF3b complex | | Descriptor: | ATP-dependent RNA helicase DDX42, PHD finger-like domain-containing protein 5A, Splicing factor 3B subunit 1, ... | | Authors: | Zhang, X, Zhan, X, Shi, Y. | | Deposit date: | 2021-05-21 | | Release date: | 2022-08-03 | | Last modified: | 2024-01-17 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structural insights into branch site proofreading by human spliceosome
Nat.Struct.Mol.Biol., 2024
|
|
7EVO
 
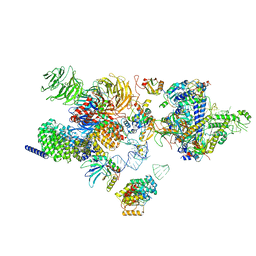 | | The cryo-EM structure of the human 17S U2 snRNP | | Descriptor: | HIV Tat-specific factor 1, PHD finger-like domain-containing protein 5A, RNA helicase, ... | | Authors: | Zhang, X, Zhan, X, Shi, Y. | | Deposit date: | 2021-05-21 | | Release date: | 2022-08-03 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Structural insights into branch site proofreading by human spliceosome.
Nat.Struct.Mol.Biol., 2024
|
|
7JVB
 
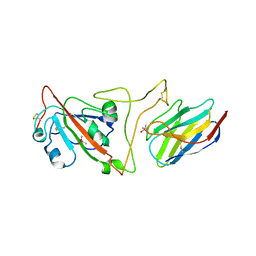 | | Crystal structure of the SARS-CoV-2 spike receptor-binding domain (RBD) with nanobody Nb20 | | Descriptor: | CACODYLATE ION, Nanobody Nb20, Spike protein S1 | | Authors: | Xiang, Y, Xiao, Z, Liu, H, Sang, Z, Schneidman-Duhovny, D, Zhang, C, Shi, Y. | | Deposit date: | 2020-08-20 | | Release date: | 2020-12-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.287 Å) | | Cite: | Versatile and multivalent nanobodies efficiently neutralize SARS-CoV-2.
Science, 370, 2020
|
|
6A8V
 
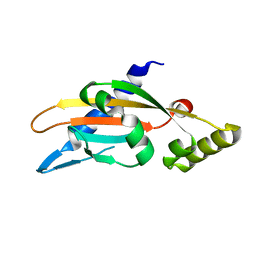 | | PhoQ sensor domain (D179R mutant): analysis of internal cavity | | Descriptor: | Sensor protein PhoQ | | Authors: | Yoshitani, K, Ishii, E, Taniguchi, K, Sugimoto, H, Shiro, Y, Mori, H, Akiyama, Y, Kato, A, Utsumi, R, Eguchi, Y. | | Deposit date: | 2018-07-10 | | Release date: | 2019-01-30 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Identification of an internal cavity in the PhoQ sensor domain for PhoQ activity and SafA-mediated control.
Biosci. Biotechnol. Biochem., 83, 2019
|
|
5ZV2
 
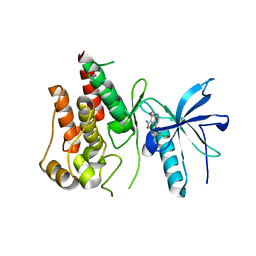 | | FGFR-1 in complex with ligand lenvatinib | | Descriptor: | 4-{3-chloro-4-[(cyclopropylcarbamoyl)amino]phenoxy}-7-methoxyquinoline-6-carboxamide, Fibroblast growth factor receptor 1 | | Authors: | Matsuki, M, Hoshi, T, Yamamoto, Y, Ikemori-Kawada, M, Minoshima, Y, Funahashi, Y, Matsui, J. | | Deposit date: | 2018-05-09 | | Release date: | 2018-07-11 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.86 Å) | | Cite: | Lenvatinib inhibits angiogenesis and tumor fibroblast growth factor signaling pathways in human hepatocellular carcinoma models.
Cancer Med, 7, 2018
|
|
1CP8
 
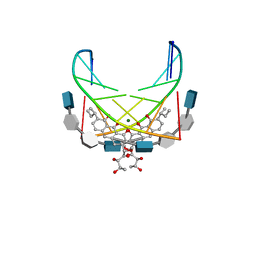 | | NMR STRUCTURE OF DNA (5'-D(TTGGCCAA)2-3') COMPLEXED WITH NOVEL ANTITUMOR DRUG UCH9 | | Descriptor: | 1,2-HYDRO-1-OXY-3,4-HYDRO-3-(1-METHOXY-2-OXY-3,4-DIHYDROXYPENTYL)-8,9-DIHYDROXY-7-(SEC-BUTYL)-ANTHRACENE, DNA (5'-D(P*TP*TP*GP*GP*CP*CP*AP*A)-3'), MAGNESIUM ION, ... | | Authors: | Katahira, R, Katahira, M, Yamashita, Y, Ogawa, H, Kyogoku, Y, Yoshida, M. | | Deposit date: | 1999-06-11 | | Release date: | 1999-07-01 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the novel antitumor drug UCH9 complexed with d(TTGGCCAA)2 as determined by NMR.
Nucleic Acids Res., 26, 1998
|
|
5Y5M
 
 | | SFX structure of cytochrome P450nor: a complete dark data without pump laser (resting state) | | Descriptor: | NADP nitrous oxide-forming nitric oxide reductase, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Tosha, T, Nomura, T, Nishida, T, Saeki, N, Okubayashi, K, Yamagiwa, R, Sugahara, M, Nakane, T, Yamashita, K, Hirata, K, Ueno, G, Kimura, T, Hisano, T, Muramoto, K, Sawai, H, Takeda, H, Mizohata, E, Yamashita, A, Kanematsu, Y, Takano, Y, Nango, E, Tanaka, R, Nureki, O, Ikemoto, Y, Murakami, H, Owada, S, Tono, K, Yabashi, M, Yamamoto, M, Ago, H, Iwata, S, Sugimoto, H, Shiro, Y, Kubo, M. | | Deposit date: | 2017-08-09 | | Release date: | 2018-08-08 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Capturing an initial intermediate during the P450nor enzymatic reaction using time-resolved XFEL crystallography and caged-substrate.
Nat Commun, 8, 2017
|
|
1JF6
 
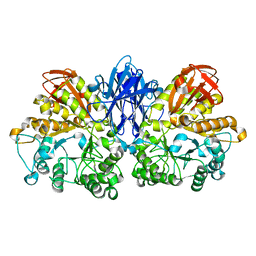 | | Crystal structure of thermoactinomyces vulgaris r-47 alpha-amylase mutant F286Y | | Descriptor: | ALPHA AMYLASE II, CALCIUM ION | | Authors: | Ohtaki, A, Kondo, S, Shimura, Y, Tonozuka, T, Sakano, Y, Kamitori, S. | | Deposit date: | 2001-06-20 | | Release date: | 2002-05-22 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Role of Phe286 in the recognition mechanism of cyclomaltooligosaccharides (cyclodextrins) by Thermoactinomyces vulgaris R-47 alpha-amylase 2 (TVAII). X-ray structures of the mutant TVAIIs, F286A and F286Y, and kinetic analyses of the Phe286-replaced mutant TVAIIs
CARBOHYDR.RES., 334, 2001
|
|
6A8U
 
 | | PhoQ sensor domain (wild type): analysis of internal cavity | | Descriptor: | Sensor protein PhoQ | | Authors: | Yoshitani, K, Ishii, E, Taniguchi, K, Sugimoto, H, Shiro, Y, Mori, H, Akiyama, Y, Kato, A, Utsumi, R, Eguchi, Y. | | Deposit date: | 2018-07-10 | | Release date: | 2019-01-30 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.848 Å) | | Cite: | Identification of an internal cavity in the PhoQ sensor domain for PhoQ activity and SafA-mediated control.
Biosci. Biotechnol. Biochem., 83, 2019
|
|
5Y5I
 
 | | Time-resolved SFX structure of cytochrome P450nor: 20 ms after photo-irradiation of caged NO in the presence of NADH (NO-bound state), light data | | Descriptor: | GLYCEROL, NADP nitrous oxide-forming nitric oxide reductase, NITRIC OXIDE, ... | | Authors: | Tosha, T, Nomura, T, Nishida, T, Saeki, N, Okubayashi, K, Yamagiwa, R, Sugahara, M, Nakane, T, Yamashita, K, Hirata, K, Ueno, G, Kimura, T, Hisano, T, Muramoto, K, Sawai, H, Takeda, H, Mizohata, E, Yamashita, A, Kanematsu, Y, Takano, Y, Nango, E, Tanaka, R, Nureki, O, Ikemoto, Y, Murakami, H, Owada, S, Tono, K, Yabashi, M, Yamamoto, M, Ago, H, Iwata, S, Sugimoto, H, Shiro, Y, Kubo, M. | | Deposit date: | 2017-08-09 | | Release date: | 2017-12-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Capturing an initial intermediate during the P450nor enzymatic reaction using time-resolved XFEL crystallography and caged-substrate.
Nat Commun, 8, 2017
|
|
5Y5L
 
 | | Time-resolved SFX structure of cytochrome P450nor: dark-2 data in the absence of NADH (resting state) | | Descriptor: | NADP nitrous oxide-forming nitric oxide reductase, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Tosha, T, Nomura, T, Nishida, T, Saeki, N, Okubayashi, K, Yamagiwa, R, Sugahara, M, Nakane, T, Yamashita, K, Hirata, K, Ueno, G, Kimura, T, Hisano, T, Muramoto, K, Sawai, H, Takeda, H, Mizohata, E, Yamashita, A, Kanematsu, Y, Takano, Y, Nango, E, Tanaka, R, Nureki, O, Ikemoto, Y, Murakami, H, Owada, S, Tono, K, Yabashi, M, Yamamoto, M, Ago, H, Iwata, S, Sugimoto, H, Shiro, Y, Kubo, M. | | Deposit date: | 2017-08-09 | | Release date: | 2017-12-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Capturing an initial intermediate during the P450nor enzymatic reaction using time-resolved XFEL crystallography and caged-substrate.
Nat Commun, 8, 2017
|
|
5Y5J
 
 | | Time-resolved SFX structure of cytochrome P450nor: dark-2 data in the presence of NADH (resting state) | | Descriptor: | GLYCEROL, NADP nitrous oxide-forming nitric oxide reductase, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Tosha, T, Nomura, T, Nishida, T, Saeki, N, Okubayashi, K, Yamagiwa, R, Sugahara, M, Nakane, T, Yamashita, K, Hirata, K, Ueno, G, Kimura, T, Hisano, T, Muramoto, K, Sawai, H, Takeda, H, Mizohata, E, Yamashita, A, Kanematsu, Y, Takano, Y, Nango, E, Tanaka, R, Nureki, O, Ikemoto, Y, Murakami, H, Owada, S, Tono, K, Yabashi, M, Yamamoto, M, Ago, H, Iwata, S, Sugimoto, H, Shiro, Y, Kubo, M. | | Deposit date: | 2017-08-09 | | Release date: | 2017-12-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Capturing an initial intermediate during the P450nor enzymatic reaction using time-resolved XFEL crystallography and caged-substrate.
Nat Commun, 8, 2017
|
|
