3INV
 
 | | Trypanosoma cruzi Dihydrofolate Reductase-Thymidylate Synthase COMPLEXED WITH NADPH, dUMP AND C-448 ANTIFOLATE | | Descriptor: | 1-[3-(2,3-dichlorophenoxy)propoxy]-6,6-dimethyl-1,6-dihydro-1,3,5-triazine-2,4-diamine, 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, Bifunctional dihydrofolate reductase-thymidylate synthase, ... | | Authors: | Chitnumsub, P, Yuvaniyama, J, Yuthavong, Y. | | Deposit date: | 2009-08-13 | | Release date: | 2010-07-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Structural basis of antifolate inhibition of Trypanosoma cruzi Dihydrofolate Reductase-Thymidylate Synthase
To be Published
|
|
5B6X
 
 | | A three dimensional movie of structural changes in bacteriorhodopsin: structure obtained 760 ns after photoexcitation | | Descriptor: | 2,3-DI-PHYTANYL-GLYCEROL, Bacteriorhodopsin, DECANE, ... | | Authors: | Royant, A, Nango, E, Nakane, T, Tanaka, T, Arima, T, Neutze, R, Iwata, S. | | Deposit date: | 2016-06-02 | | Release date: | 2016-12-21 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A three-dimensional movie of structural changes in bacteriorhodopsin
Science, 354, 2016
|
|
3IRN
 
 | | Trypanosoma cruzi Dihydrofolate Reductase-Thymidylate Synthase COMPLEXED WITH NADPH AND Cycloguanil | | Descriptor: | 1-(4-chlorophenyl)-6,6-dimethyl-1,6-dihydro-1,3,5-triazine-2,4-diamine, Bifunctional dihydrofolate reductase-thymidylate synthase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Chitnumsub, P, Yuvaniyama, J, Yuthavong, Y. | | Deposit date: | 2009-08-24 | | Release date: | 2010-07-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis of antifolate inhibition of Trypanosoma cruzi Dihydrofolate Reductase-Thymidylate Synthase
To be Published
|
|
3ER5
 
 | | THE ACTIVE SITE OF ASPARTIC PROTEINASES | | Descriptor: | ENDOTHIAPEPSIN, H-189 | | Authors: | Bailey, D, Veerapandian, B, Cooper, J, Szelke, M, Blundell, T.L. | | Deposit date: | 1991-01-05 | | Release date: | 1991-04-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-ray-crystallographic studies of complexes of pepstatin A and a statine-containing human renin inhibitor with endothiapepsin.
Biochem.J., 289 ( Pt 2), 1993
|
|
5B6Y
 
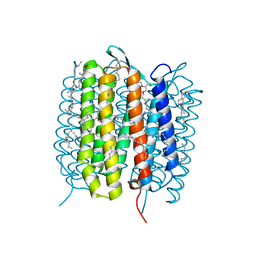 | | A three dimensional movie of structural changes in bacteriorhodopsin: structure obtained 36.2 us after photoexcitation | | Descriptor: | 2,3-DI-PHYTANYL-GLYCEROL, Bacteriorhodopsin, DECANE, ... | | Authors: | Royant, A, Nango, E, Nakane, T, Tanaka, T, Arima, T, Neutze, R, Iwata, S. | | Deposit date: | 2016-06-02 | | Release date: | 2016-12-21 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A three-dimensional movie of structural changes in bacteriorhodopsin
Science, 354, 2016
|
|
3ER3
 
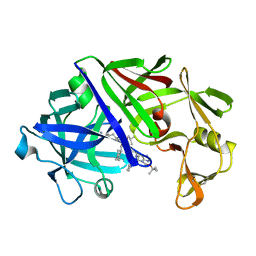 | | The active site of aspartic proteinases | | Descriptor: | 6-ammonio-N-[(2R,4R,5R)-5-{[N-(tert-butoxycarbonyl)-L-phenylalanyl-3-(1H-imidazol-3-ium-4-yl)-L-alanyl]amino}-6-cyclohexyl-4-hydroxy-2-(2-methylpropyl)hexanoyl]-L-norleucylphenylalanine, ENDOTHIAPEPSIN | | Authors: | Al-Karadaghi, S, Cooper, J.B, Veerapandian, B, Hoover, D, Blundell, T.L. | | Deposit date: | 1991-01-02 | | Release date: | 1991-04-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Active Site of Aspartic Proteinases
FEBS Lett., 174, 1984
|
|
5EDS
 
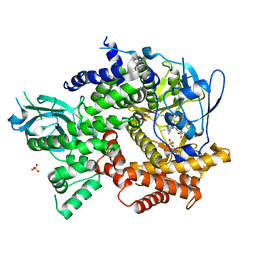 | | Crystal structure of human PI3K-gamma in complex with benzimidazole inhibitor 5 | | Descriptor: | 4-azanyl-6-[[(1~{S})-1-[6-fluoranyl-1-(3-methylsulfonylphenyl)benzimidazol-2-yl]ethyl]amino]pyrimidine-5-carbonitrile, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit gamma isoform, SULFATE ION | | Authors: | Whittington, D.A, Tang, J, Yakowec, P. | | Deposit date: | 2015-10-21 | | Release date: | 2015-12-30 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Discovery, Optimization, and in Vivo Evaluation of Benzimidazole Derivatives AM-8508 and AM-9635 as Potent and Selective PI3K delta Inhibitors.
J.Med.Chem., 59, 2016
|
|
5HYX
 
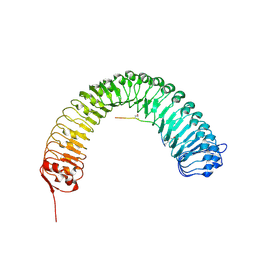 | | Plant peptide hormone receptor RGFR1 in complex with RGF1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, PTR-SER-ASN-PRO-GLY-HIS-HIS-PRO-HYP-ARG-HIS-ASN, ... | | Authors: | Song, W, Han, Z, Chai, J. | | Deposit date: | 2016-02-02 | | Release date: | 2017-01-11 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (2.596 Å) | | Cite: | Signature motif-guided identification of receptors for peptide hormones essential for root meristem growth
Cell Res., 26, 2016
|
|
8PVQ
 
 | | Crystal Structure of Human PTX3 C-terminal domain | | Descriptor: | Pentraxin-related protein PTX3 | | Authors: | Levy, C.W. | | Deposit date: | 2023-07-18 | | Release date: | 2024-07-24 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Pentraxin 3 mediated crosslinking of heavy chain hyaluronan complexes: new insights from structural and biophysical studies
To Be Published
|
|
8PWJ
 
 | | Light structure of sensory rhodopsin-II solved by serial millisecond crystallography. 30-60 milliseconds time-bin | | Descriptor: | CHLORIDE ION, RETINAL, Sensory rhodopsin-2 | | Authors: | Bosman, R, Ortolani, G, Branden, G, Neutze, R. | | Deposit date: | 2023-07-20 | | Release date: | 2025-02-05 | | Last modified: | 2025-04-23 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Structural basis for the prolonged photocycle of sensory rhodopsin II revealed by serial synchrotron crystallography.
Nat Commun, 16, 2025
|
|
8PWQ
 
 | | Light structure of sensory rhodopsin-II solved by serial millisecond crystallography 120-150 milliseconds time-bin | | Descriptor: | CHLORIDE ION, RETINAL, Sensory rhodopsin-2 | | Authors: | Bosman, R, Ortolani, G, Branden, G, Neutze, R. | | Deposit date: | 2023-07-21 | | Release date: | 2025-02-05 | | Last modified: | 2025-04-23 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural basis for the prolonged photocycle of sensory rhodopsin II revealed by serial synchrotron crystallography.
Nat Commun, 16, 2025
|
|
8PWI
 
 | | Light structure of sensory rhodopsin-II solved by serial millisecond crystallography 60-90 milliseconds time-bin | | Descriptor: | CHLORIDE ION, RETINAL, Sensory rhodopsin-2 | | Authors: | Bosman, R, Ortolani, G, Branden, G, Neutze, R. | | Deposit date: | 2023-07-20 | | Release date: | 2025-02-05 | | Last modified: | 2025-04-23 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Structural basis for the prolonged photocycle of sensory rhodopsin II revealed by serial synchrotron crystallography.
Nat Commun, 16, 2025
|
|
8PWG
 
 | | Light structure of sensory rhodopsin-II solved by serial millisecond crystallography 90-120 milliseconds time-bin | | Descriptor: | CHLORIDE ION, RETINAL, Sensory rhodopsin-2 | | Authors: | Ortolani, G, Bosman, R, Branden, G, Neutze, R. | | Deposit date: | 2023-07-20 | | Release date: | 2025-02-05 | | Last modified: | 2025-04-23 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Structural basis for the prolonged photocycle of sensory rhodopsin II revealed by serial synchrotron crystallography.
Nat Commun, 16, 2025
|
|
8YHE
 
 | | Cryo-EM structure of CTR-bound type VII CRISPR-Cas complex at post-state II | | Descriptor: | RNA (29-MER), RNA (46-MER), ZINC ION, ... | | Authors: | Zhang, H, Deng, Z, Li, X. | | Deposit date: | 2024-02-28 | | Release date: | 2024-08-21 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.07 Å) | | Cite: | Structural basis for the activity of the type VII CRISPR-Cas system.
Nature, 633, 2024
|
|
8Z4J
 
 | | Cryo-EM structure of CTR-bound type VII CRISPR-Cas complex at substrate-engaged state II | | Descriptor: | Protein structure, RNA (34-MER), RNA (38-MER), ... | | Authors: | Zhang, H, Deng, Z, Li, X. | | Deposit date: | 2024-04-17 | | Release date: | 2024-08-21 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.97 Å) | | Cite: | Structural basis for the activity of the type VII CRISPR-Cas system.
Nature, 633, 2024
|
|
8Z9C
 
 | | Cryo-EM structure of NTR-bound type VII CRISPR-Cas complex at substrate-engaged state I | | Descriptor: | Protein structure, RNA (41-MER), RNA (48-MER), ... | | Authors: | Zhang, H, Deng, Z, Li, X. | | Deposit date: | 2024-04-23 | | Release date: | 2024-08-21 | | Last modified: | 2024-11-27 | | Method: | ELECTRON MICROSCOPY (3.01 Å) | | Cite: | Structural basis for the activity of the type VII CRISPR-Cas system.
Nature, 633, 2024
|
|
8PWP
 
 | | Light structure of sensory rhodopsin-II solved by serial millisecond crystallography 0-30 milliseconds time-bin | | Descriptor: | CHLORIDE ION, RETINAL, Sensory rhodopsin-2 | | Authors: | Ortolani, G, Bosman, R, Branden, G, Neutze, R. | | Deposit date: | 2023-07-21 | | Release date: | 2025-02-05 | | Last modified: | 2025-04-23 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Structural basis for the prolonged photocycle of sensory rhodopsin II revealed by serial synchrotron crystallography.
Nat Commun, 16, 2025
|
|
8Z9E
 
 | | Cryo-EM structure of NTR-bound type VII CRISPR-Cas complex at substrate-engaged state II | | Descriptor: | Protein structure, RNA (34-MER), RNA (39-MER), ... | | Authors: | Zhang, H, Deng, Z, Li, X. | | Deposit date: | 2024-04-23 | | Release date: | 2024-08-21 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.13 Å) | | Cite: | Structural basis for the activity of the type VII CRISPR-Cas system.
Nature, 633, 2024
|
|
8Z4L
 
 | | Cryo-EM structure of CTR-bound type VII CRISPR-Cas complex at substrate-engaged state I | | Descriptor: | RNA (40-MER), RNA (49-MER), ZINC ION, ... | | Authors: | Zhang, H, Deng, Z, Li, X. | | Deposit date: | 2024-04-17 | | Release date: | 2024-08-21 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.85 Å) | | Cite: | Structural basis for the activity of the type VII CRISPR-Cas system.
Nature, 633, 2024
|
|
8Z99
 
 | | Cryo-EM structure of NTR-bound type VII CRISPR-Cas complex at substrate-engaged state +I | | Descriptor: | RNA (49-MER), RNA (54-MER), ZINC ION, ... | | Authors: | Zhang, H, Deng, Z, Li, X. | | Deposit date: | 2024-04-22 | | Release date: | 2024-08-21 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural basis for the activity of the type VII CRISPR-Cas system.
Nature, 633, 2024
|
|
8YHD
 
 | | Cryo-EM structure of CTR-bound type VII CRISPR-Cas complex at post-state I | | Descriptor: | RNA (35-MER), RNA (53-MER), ZINC ION, ... | | Authors: | Zhang, H, Deng, Z, Li, X. | | Deposit date: | 2024-02-28 | | Release date: | 2024-08-21 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (2.93 Å) | | Cite: | Structural basis for the activity of the type VII CRISPR-Cas system.
Nature, 633, 2024
|
|
6HF5
 
 | | Crystal Structure of the Acquired VIM-2 Metallo-beta-Lactamase in Complex with ANT-431 Inhibitor | | Descriptor: | 5-(pyridin-3-ylsulfonylamino)-1,3-thiazole-4-carboxylic acid, ACETATE ION, Beta-lactamase class B VIM-2, ... | | Authors: | Docquier, J.D, Pozzi, C, De Luca, F, Benvenuti, M, Mangani, S. | | Deposit date: | 2018-08-21 | | Release date: | 2018-12-05 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | SAR Studies Leading to the Identification of a Novel Series of Metallo-beta-lactamase Inhibitors for the Treatment of Carbapenem-Resistant Enterobacteriaceae Infections That Display Efficacy in an Animal Infection Model.
Acs Infect Dis., 5, 2019
|
|
8QV6
 
 | | Structure of human SPNS2 in DDM | | Descriptor: | DODECYL-BETA-D-MALTOSIDE, Nanobody D12, Sphingosine-1-phosphate transporter SPNS2 | | Authors: | Li, H.Z, Pike, A.C.W, McKinley, G, Mukhopadhyay, S.M.M, Moreau, C, Scacioc, A, Abrusci, P, Borkowska, O, Chalk, R, Stefanic, S, Burgess-Brown, N, Duerr, K.L, Sauer, D.B. | | Deposit date: | 2023-10-17 | | Release date: | 2025-02-19 | | Method: | ELECTRON MICROSCOPY (3.68 Å) | | Cite: | Transport and inhibition of the sphingosine-1-phosphate exporter SPNS2.
Nat Commun, 16, 2025
|
|
8QV5
 
 | | Structure of human SPNS2 in LMNG | | Descriptor: | Nanobody D12, Sphingosine-1-phosphate transporter SPNS2 | | Authors: | Li, H.Z, Pike, A.C.W, McKinley, G, Mukhopadhyay, S.M.M, Moreau, C, Scacioc, A, Abrusci, P, Borkowska, O, Chalk, R, Stefanic, S, Burgess-Brown, N, Duerr, K.L, Sauer, D.B. | | Deposit date: | 2023-10-17 | | Release date: | 2025-02-19 | | Method: | ELECTRON MICROSCOPY (3.69 Å) | | Cite: | Transport and inhibition of the sphingosine-1-phosphate exporter SPNS2.
Nat Commun, 16, 2025
|
|
7W1Q
 
 | |
