3L82
 
 | | X-ray Crystal structure of TRF1 and Fbx4 complex | | Descriptor: | F-box only protein 4, Telomeric repeat-binding factor 1 | | Authors: | Zeng, Z.X, Wang, W, Yang, Y.T, Chen, Y, Yang, X.M, Diehl, J.A, Liu, X.D, Lei, M. | | Deposit date: | 2009-12-29 | | Release date: | 2010-03-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Basis of Selective Ubiquitination of TRF1 by SCF(Fbx4)
Dev.Cell, 18, 2010
|
|
7QXI
 
 | |
7QWP
 
 | | CryoEM structure of bacterial transcription close complex (RPc) | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Ye, F.Z, Zhang, X.D. | | Deposit date: | 2022-01-25 | | Release date: | 2022-11-09 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Mechanisms of DNA opening revealed in AAA+ transcription complex structures.
Sci Adv, 8, 2022
|
|
7QV9
 
 | |
6LP8
 
 | | Crystal structure of human DHODH in complex with inhibitor 1243 | | Descriptor: | 3-[4-[3-(dimethylamino)phenyl]-3,5-bis(fluoranyl)phenyl]benzo[f]benzotriazole-4,9-dione, ACETATE ION, Dihydroorotate dehydrogenase (quinone), ... | | Authors: | Chen, Q, Yu, Y. | | Deposit date: | 2020-01-09 | | Release date: | 2020-09-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Bifunctional Naphtho[2,3- d ][1,2,3]triazole-4,9-dione Compounds Exhibit Antitumor Effects In Vitro and In Vivo by Inhibiting Dihydroorotate Dehydrogenase and Inducing Reactive Oxygen Species Production.
J.Med.Chem., 63, 2020
|
|
5XJN
 
 | | cytochrome P450 CREJ in complex with (4-ethylphenyl) dihydrogen phosphate | | Descriptor: | (4-ethylphenyl) dihydrogen phosphate, Cytochrome P450, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Dong, S, Du, L, Li, S, Feng, Y. | | Deposit date: | 2017-05-03 | | Release date: | 2017-07-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Selective oxidation of aliphatic C-H bonds in alkylphenols by a chemomimetic biocatalytic system
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
3WDO
 
 | | Structure of E. coli YajR transporter | | Descriptor: | MFS Transporter | | Authors: | Jiang, D. | | Deposit date: | 2013-06-19 | | Release date: | 2013-08-07 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Structure of the YajR transporter suggests a transport mechanism based on the conserved motif A
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
7YE9
 
 | | SARS-CoV-2 Spike (6P) in complex with 3 R1-32 Fabs | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of R1-32 Fab, ... | | Authors: | Liu, B, Gao, X, Li, Z, Chen, X, He, J, Chen, L, Xiong, X. | | Deposit date: | 2022-07-05 | | Release date: | 2022-08-24 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (4.17 Å) | | Cite: | SARS-CoV-2 Delta and Omicron variants evade population antibody response by mutations in a single spike epitope.
Nat Microbiol, 7, 2022
|
|
9JEA
 
 | |
7YDY
 
 | | SARS-CoV-2 Spike (6P) in complex with 1 R1-32 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of R1-32 Fab, ... | | Authors: | Liu, B, Gao, X, Li, Z, Chen, X, He, J, Chen, L, Xiong, X. | | Deposit date: | 2022-07-04 | | Release date: | 2022-08-24 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (4.75 Å) | | Cite: | SARS-CoV-2 Delta and Omicron variants evade population antibody response by mutations in a single spike epitope.
Nat Microbiol, 7, 2022
|
|
7YEG
 
 | | SARS-CoV-2 Spike (6P) in complex with 3 R1-32 Fabs and 3 ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Liu, B, Gao, X, Li, Z, Chen, X, He, J, Chen, L, Xiong, X. | | Deposit date: | 2022-07-05 | | Release date: | 2022-08-24 | | Last modified: | 2025-03-12 | | Method: | ELECTRON MICROSCOPY (3.73 Å) | | Cite: | SARS-CoV-2 Delta and Omicron variants evade population antibody response by mutations in a single spike epitope.
Nat Microbiol, 7, 2022
|
|
7YDI
 
 | | SARS-CoV-2 Spike (6P) in complex with 3 R1-32 Fabs and 3 ACE2, focused refinement of RBD region | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of R1-32, Light chain of R1-32, ... | | Authors: | Liu, B, Gao, X, Li, Z, Chen, X, He, J, Chen, L, Xiong, X. | | Deposit date: | 2022-07-04 | | Release date: | 2022-08-24 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.98 Å) | | Cite: | SARS-CoV-2 Delta and Omicron variants evade population antibody response by mutations in a single spike epitope.
Nat Microbiol, 7, 2022
|
|
7YE5
 
 | | SARS-CoV-2 Spike (6P) in complex with 2 R1-32 Fabs | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of R1-32 Fab, ... | | Authors: | Liu, B, Gao, X, Li, Z, Chen, X, He, J, Chen, L, Xiong, X. | | Deposit date: | 2022-07-05 | | Release date: | 2022-08-24 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (6.75 Å) | | Cite: | SARS-CoV-2 Delta and Omicron variants evade population antibody response by mutations in a single spike epitope.
Nat Microbiol, 7, 2022
|
|
7XP6
 
 | | Cryo-EM structure of a class T GPCR in active state | | Descriptor: | Endoglucanase H,Taste receptor type 2 member 46,Endoglucanase H,Taste receptor type 2 member 46,Bitter taste receptor T2R46, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Liu, Z.J, Hua, T, Xu, W.X, Wu, L.J. | | Deposit date: | 2022-05-03 | | Release date: | 2022-10-12 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.01 Å) | | Cite: | Structural basis for strychnine activation of human bitter taste receptor TAS2R46.
Science, 377, 2022
|
|
7XP4
 
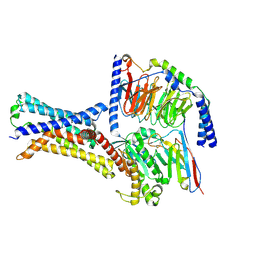 | | Cryo-EM structure of a class T GPCR in apo state | | Descriptor: | Endoglucanase H,Taste receptor type 2 member 46,Endoglucanase H,Taste receptor type 2 member 46,Bitter taste receptor T2R46, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Liu, Z.J, Hua, T, Xu, W.X, Wu, L.J. | | Deposit date: | 2022-05-03 | | Release date: | 2022-10-12 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.01 Å) | | Cite: | Structural basis for strychnine activation of human bitter taste receptor TAS2R46.
Science, 377, 2022
|
|
7XP5
 
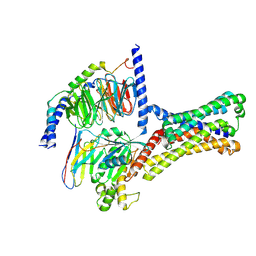 | | Cryo-EM structure of a class T GPCR in ligand-free state | | Descriptor: | Endoglucanase H,Taste receptor type 2 member 46,Bitter taste receptor T2R46, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Liu, Z.J, Hua, T, Xu, W.X, Wu, L.J. | | Deposit date: | 2022-05-03 | | Release date: | 2022-10-12 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.08 Å) | | Cite: | Structural basis for strychnine activation of human bitter taste receptor TAS2R46.
Science, 377, 2022
|
|
7KDT
 
 | |
1KVW
 
 | |
1KVY
 
 | |
9LX5
 
 | | Cryo-EM structure of the P2X1 receptor bound to ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CHOLESTEROL, P2X purinoceptor 1 | | Authors: | Qiang, Y, Chen, K. | | Deposit date: | 2025-02-17 | | Release date: | 2025-04-16 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural basis of the multiple ligand binding mechanisms of the P2X1 receptor.
Acta Pharmacol.Sin., 2025
|
|
9LXC
 
 | | Cryo-EM structure of the P2X1 receptor bound to NF449 | | Descriptor: | 4,4',4'',4'''-{carbonylbis[azanediylbenzene-5,1,3-triylbis(carbonylazanediyl)]}tetra(benzene-1,3-disulfonic acid), CHOLESTEROL, P2X purinoceptor 1 | | Authors: | Qiang, Y, Chen, K. | | Deposit date: | 2025-02-17 | | Release date: | 2025-04-16 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis of the multiple ligand binding mechanisms of the P2X1 receptor.
Acta Pharmacol.Sin., 2025
|
|
3TB4
 
 | | Crystal structure of the ISC domain of VibB | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Liu, S, Zhang, C, Niu, B, Li, N, Liu, M, Wei, T, Zhu, D, Xu, S, Gu, L. | | Deposit date: | 2011-08-05 | | Release date: | 2012-08-29 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structural insight into the ISC domain of VibB from Vibrio cholerae at atomic resolution: a snapshot just before the enzymatic reaction
Acta Crystallogr.,Sect.D, 68, 2012
|
|
4Z9P
 
 | | Crystal structure of Ebola virus nucleoprotein core domain at 1.8A resolution | | Descriptor: | Nucleoprotein | | Authors: | Guo, Y, Dong, S.S, Yang, P, Li, G.B, Liu, B.C, Yang, C, Rao, Z.H. | | Deposit date: | 2015-04-11 | | Release date: | 2015-05-20 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.792 Å) | | Cite: | Insight into the Ebola virus nucleocapsid assembly mechanism: crystal structure of Ebola virus nucleoprotein core domain at 1.8 A resolution.
Protein Cell, 6, 2015
|
|
9DC9
 
 | | Mtb GuaB dCBS in complex with inhibitor compound 5 | | Descriptor: | (3P)-3-(4-chloro-1H-imidazol-2-yl)-5-[(1R)-1-{[(3M)-3-(1H-pyrazol-5-yl)phenyl]oxy}ethyl]pyridine, INOSINIC ACID, Inosine-5'-monophosphate dehydrogenase | | Authors: | Harris, S.F, Wu, P. | | Deposit date: | 2024-08-25 | | Release date: | 2024-11-27 | | Last modified: | 2025-01-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Discovery of potent dihydro-oxazinoquinolinone inhibitors of GuaB for the treatment of tuberculosis.
Bioorg.Med.Chem.Lett., 117, 2025
|
|
9DC8
 
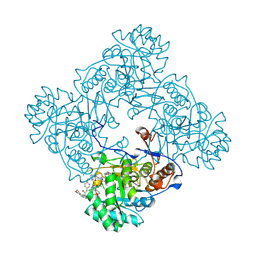 | | Mtb GuaB dCBS in complex with inhibitor G1 | | Descriptor: | INOSINIC ACID, Inosine-5'-monophosphate dehydrogenase, N-(6-chloropyridin-3-yl)-N~2~-(1,4-dihydro-2H-pyrano[3,4-c]quinolin-9-yl)-L-alaninamide, ... | | Authors: | Harris, S.F, Wu, P. | | Deposit date: | 2024-08-25 | | Release date: | 2024-11-27 | | Last modified: | 2025-01-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Discovery of potent dihydro-oxazinoquinolinone inhibitors of GuaB for the treatment of tuberculosis.
Bioorg.Med.Chem.Lett., 117, 2025
|
|
