3MIP
 
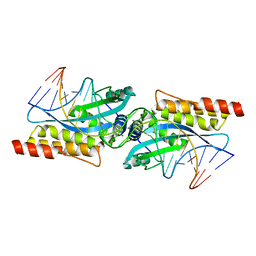 | | I-MsoI re-designed for altered DNA cleavage specificity (-8GCG) | | Descriptor: | CALCIUM ION, DNA (5'-D(*CP*GP*GP*AP*GP*CP*GP*GP*TP*CP*TP*CP*AP*CP*GP*AP*CP*CP*GP*CP*CP*TP*GP*C)-3'), DNA (5'-D(*GP*CP*AP*GP*GP*CP*GP*GP*TP*CP*GP*TP*GP*AP*GP*AP*CP*CP*GP*CP*TP*CP*CP*G)-3'), ... | | Authors: | Taylor, G.K, Stoddard, B.L. | | Deposit date: | 2010-04-11 | | Release date: | 2010-05-19 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Computational reprogramming of homing endonuclease specificity at multiple adjacent base pairs.
Nucleic Acids Res., 38, 2010
|
|
3MIS
 
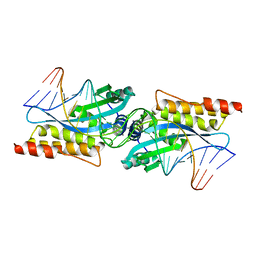 | | I-MsoI re-designed for altered DNA cleavage specificity (-8G) | | Descriptor: | CALCIUM ION, DNA (5'-D(*CP*GP*GP*AP*GP*CP*TP*GP*TP*CP*TP*CP*AP*CP*GP*AP*CP*GP*TP*CP*CP*TP*GP*C)-3'), DNA (5'-D(*GP*CP*AP*GP*GP*AP*CP*GP*TP*CP*GP*TP*GP*AP*GP*AP*CP*AP*GP*CP*TP*CP*CP*G)-3'), ... | | Authors: | Taylor, G.K, Stoddard, B.L. | | Deposit date: | 2010-04-12 | | Release date: | 2010-05-19 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Computational reprogramming of homing endonuclease specificity at multiple adjacent base pairs.
Nucleic Acids Res., 38, 2010
|
|
2LNU
 
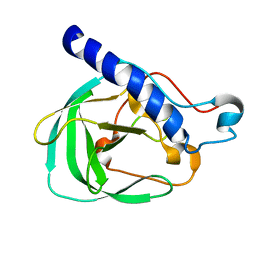 | | Solution NMR Structure of the uncharacterized protein from gene locus rrnAC0354 of Haloarcula marismortui, Northeast Structural Genomics Consortium Target HmR11 | | Descriptor: | Uncharacterized protein | | Authors: | Rossi, P, Liu, G, Lange, O.F, Lee, H, Janjua, H, Ciccosanti, C, Wang, H, Acton, T.B, Xiao, R, Everett, J.K, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2012-01-05 | | Release date: | 2012-01-18 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Determination of solution structures of proteins up to 40 kDa using CS-Rosetta with sparse NMR data from deuterated samples.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
2LOK
 
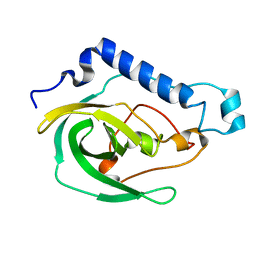 | | Solution NMR Structure of the uncharacterized protein from gene locus VNG_0733H of Halobacterium salinarium, Northeast Structural Genomics Consortium Target HsR50 | | Descriptor: | Uncharacterized protein | | Authors: | Rossi, P, Lange, O.F, Lee, H, Hamilton, K, Ciccosanti, C, Buchwald, W.A, Wang, H, Acton, T.B, Xiao, R, Everett, J.K, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2012-01-24 | | Release date: | 2012-02-01 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Determination of solution structures of proteins up to 40 kDa using CS-Rosetta with sparse NMR data from deuterated samples.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
2KZN
 
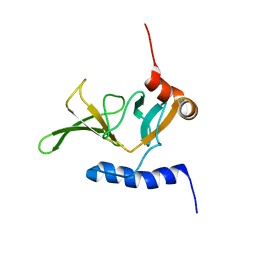 | | Solution NMR Structure of Peptide methionine sulfoxide reductase msrB from Bacillus subtilis, Northeast Structural Genomics Consortium Target SR10 | | Descriptor: | Peptide methionine sulfoxide reductase msrB | | Authors: | Ertekin, A, Maglaqui, M, Janjua, H, Cooper, B, Ciccosanti, C, Rost, B, Acton, T.B, Xiao, R, Everett, J.K, Prestegard, J, Lee, H, Aramini, J.M, Rossi, P, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2010-06-18 | | Release date: | 2010-07-07 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Determination of solution structures of proteins up to 40 kDa using CS-Rosetta with sparse NMR data from deuterated samples.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4J3O
 
 | |
4OYC
 
 | |
4JLR
 
 | | Crystal structure of a designed Respiratory Syncytial Virus Immunogen in complex with Motavizumab | | Descriptor: | Motavizumab Fab heavy chain, Motavizumab Fab light chain, PENTAETHYLENE GLYCOL, ... | | Authors: | Rupert, P.B, Correia, B, Schief, W, Strong, R.K. | | Deposit date: | 2013-03-12 | | Release date: | 2014-02-05 | | Last modified: | 2014-12-17 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Proof of principle for epitope-focused vaccine design.
Nature, 507, 2014
|
|
4OU1
 
 | | Crystal structure of a computationally designed retro-aldolase covalently bound to folding probe 1 [(6-methoxynaphthalen-2-yl)(oxiran-2-yl)methanol] | | Descriptor: | (1S,2S)-1-(6-methoxynaphthalen-2-yl)propane-1,2-diol, BENZOIC ACID, PHOSPHATE ION, ... | | Authors: | Bhabha, G, Zhang, X, Ekiert, D.C. | | Deposit date: | 2014-02-14 | | Release date: | 2014-03-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Small molecule probes to quantify the functional fraction of a specific protein in a cell with minimal folding equilibrium shifts.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4L8I
 
 | | Crystal structure of RSV epitope scaffold FFL_005 | | Descriptor: | RSV epitope scaffold FFL_005 | | Authors: | Jardine, J, Correnti, C, Holmes, M.A, Strong, R.K, Schief, W.R. | | Deposit date: | 2013-06-17 | | Release date: | 2014-02-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Proof of principle for epitope-focused vaccine design.
Nature, 507, 2014
|
|
4R6J
 
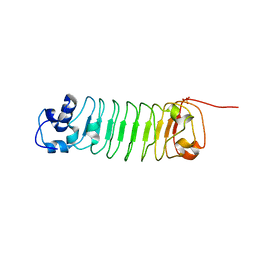 | |
4J8T
 
 | | Engineered Digoxigenin binder DIG10.2 | | Descriptor: | DIGOXIGENIN, Engineered Digoxigenin binder protein DIG10.2 | | Authors: | Stoddard, B.L, Doyle, L.A. | | Deposit date: | 2013-02-14 | | Release date: | 2013-06-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Computational design of ligand-binding proteins with high affinity and selectivity.
Nature, 501, 2013
|
|
4J9A
 
 | | Engineered Digoxigenin binder DIG10.3 | | Descriptor: | DIGOXIGENIN, Engineered Digoxigenin binder protein DIG10.3 | | Authors: | Stoddard, B.L, Doyle, L.A. | | Deposit date: | 2013-02-15 | | Release date: | 2013-06-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Computational design of ligand-binding proteins with high affinity and selectivity.
Nature, 501, 2013
|
|
4PEJ
 
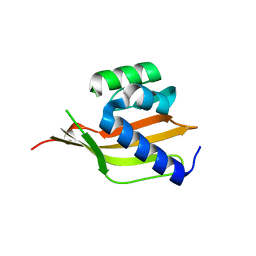 | | Crystal structure of a computationally designed retro-aldolase, RA110.4 (Cys free) | | Descriptor: | Retro-aldolase | | Authors: | Bhabha, G, Zhang, X, Liu, Y, Ekiert, D.C. | | Deposit date: | 2014-04-23 | | Release date: | 2015-04-08 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | De novo-designed enzymes as small-molecule-regulated fluorescence imaging tags and fluorescent reporters.
J.Am.Chem.Soc., 136, 2014
|
|
4R6G
 
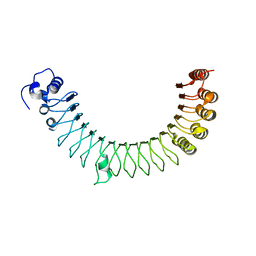 | |
4PEK
 
 | | Crystal structure of a computationally designed retro-aldolase, RA114.3 | | Descriptor: | Retro-aldolase | | Authors: | Bhabha, G, Zhang, X, Liu, Y, Ekiert, D.C. | | Deposit date: | 2014-04-23 | | Release date: | 2015-04-08 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | De novo-designed enzymes as small-molecule-regulated fluorescence imaging tags and fluorescent reporters.
J.Am.Chem.Soc., 136, 2014
|
|
4R5D
 
 | |
4R58
 
 | |
4R5C
 
 | |
4QQ8
 
 | | Crystal structure of the formolase FLS in space group P 43 21 2 | | Descriptor: | 1,2-ETHANEDIOL, Formolase, MAGNESIUM ION, ... | | Authors: | Shen, B.W, Siegel, J.B, Stoddard, B.L. | | Deposit date: | 2014-06-26 | | Release date: | 2015-03-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.88 Å) | | Cite: | Computational protein design enables a novel one-carbon assimilation pathway.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4R6F
 
 | |
2LMD
 
 | | Minimal Constraints Solution NMR Structure of Prospero Homeobox protein 1 from Homo sapiens, Northeast Structural Genomics Consortium Target HR4660B | | Descriptor: | Prospero homeobox protein 1 | | Authors: | Rossi, P, Lange, O.A, Lee, H, Maglaqui, M, Janjua, H, Ciccosanti, C, Zhao, L, Acton, T.B, Xiao, R, Everett, J.K, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2011-11-29 | | Release date: | 2011-12-21 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Determination of solution structures of proteins up to 40 kDa using CS-Rosetta with sparse NMR data from deuterated samples.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
2LOY
 
 | | Refined Miminal Constraint Solution NMR Structure of Translationally-controlled tumor protein (TCTP) from Caenorhabditis elegans, Northeast Structural Genomics Consortium Target WR73 | | Descriptor: | Translationally-controlled tumor protein homolog | | Authors: | Aramini, J.M, Rossi, P, Cort, J.R, Lee, H, Janjua, H, Maglaqui, M, Cooper, B, Xiao, R, Acton, T.B, Everett, J.K, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2012-01-27 | | Release date: | 2012-02-15 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Determination of solution structures of proteins up to 40 kDa using CS-Rosetta with sparse NMR data from deuterated samples.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
2MKY
 
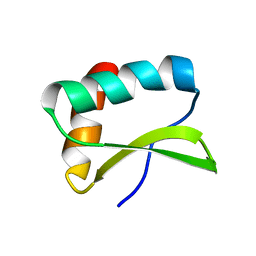 | |
3RPT
 
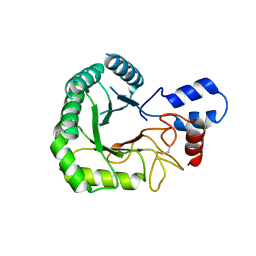 | |
