4IK3
 
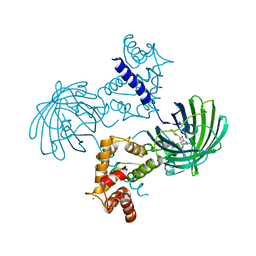 | | High resolution structure of GCaMP3 at pH 8.5 | | Descriptor: | CALCIUM ION, RCaMP, Green fluorescent protein | | Authors: | Chen, Y, Song, X, Miao, L, Zhu, Y, Ji, G. | | Deposit date: | 2012-12-25 | | Release date: | 2014-01-29 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.007 Å) | | Cite: | Structural insight into enhanced calcium indicator GCaMP3 and GCaMPJ to promote further improvement.
Protein Cell, 4, 2013
|
|
4IK9
 
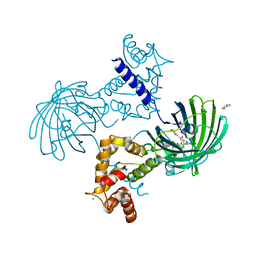 | | High resolution structure of GCaMP3 dimer form 2 at pH 7.5 | | Descriptor: | CALCIUM ION, DI(HYDROXYETHYL)ETHER, RCaMP, ... | | Authors: | Chen, Y, Song, X, Miao, L, Zhu, Y, Ji, G. | | Deposit date: | 2012-12-25 | | Release date: | 2014-01-29 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural insight into enhanced calcium indicator GCaMP3 and GCaMPJ to promote further improvement.
Protein Cell, 4, 2013
|
|
4IK5
 
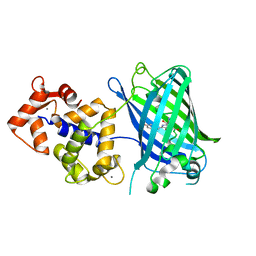 | | High resolution structure of Delta-REST-GCaMP3 | | Descriptor: | CALCIUM ION, RCaMP, Green fluorescent protein | | Authors: | Chen, Y, Song, X, Miao, L, Zhu, Y, Ji, G. | | Deposit date: | 2012-12-25 | | Release date: | 2014-01-29 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural insight into enhanced calcium indicator GCaMP3 and GCaMPJ to promote further improvement.
Protein Cell, 4, 2013
|
|
4IK1
 
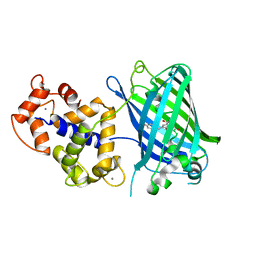 | | High resolution structure of GCaMPJ at pH 8.5 | | Descriptor: | CALCIUM ION, RCaMP, Green fluorescent protein | | Authors: | Chen, Y, Song, X, Miao, L, Zhu, Y, Ji, G. | | Deposit date: | 2012-12-25 | | Release date: | 2014-02-05 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural insight into enhanced calcium indicator GCaMP3 and GCaMPJ to promote further improvement.
Protein Cell, 4, 2013
|
|
4IK4
 
 | | High resolution structure of GCaMP3 at pH 5.0 | | Descriptor: | CALCIUM ION, RCaMP, Green fluorescent protein | | Authors: | Chen, Y, Song, X, Miao, L, Zhu, Y, Ji, G. | | Deposit date: | 2012-12-25 | | Release date: | 2014-02-05 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Structural insight into enhanced calcium indicator GCaMP3 and GCaMPJ to promote further improvement.
Protein Cell, 4, 2013
|
|
7WJR
 
 | |
4I1L
 
 | | Structural and Biological Features of FOXP3 Dimerization Relevant to Regulatory T Cell Function | | Descriptor: | ACETATE ION, Forkhead box protein P3, MAGNESIUM ION, ... | | Authors: | Song, X.M, Greene, M.I, Zhou, Z.C. | | Deposit date: | 2012-11-21 | | Release date: | 2012-12-05 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural and biological features of FOXP3 dimerization relevant to regulatory T cell function.
Cell Rep, 1, 2012
|
|
6IH5
 
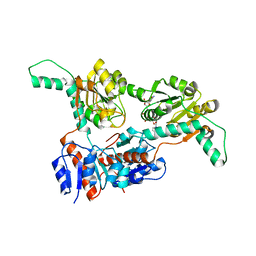 | | Crystal structure of Phosphite Dehydrogenase mutant I151R/P176E from Ralstonia sp. 4506 in complex with non-natural cofactor Nicotinamide Cytosine dinucleotide | | Descriptor: | Phosphite dehydrogenase, [[(2S,3S,4R,5S)-5-(3-aminocarbonylpyridin-1-ium-1-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl] [(2S,3S,4R,5S)-5-(4-azanyl-2-oxidanylidene-pyrimidin-1-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methyl hydrogen phosphate | | Authors: | Song, X, Feng, Y, Liu, Y, Zhao, Z. | | Deposit date: | 2018-09-28 | | Release date: | 2019-03-13 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.468 Å) | | Cite: | Structural Insights into Phosphite Dehydrogenase Variants Favoring a Non-natural Redox Cofactor
Acs Catalysis, 9, 2019
|
|
6IH4
 
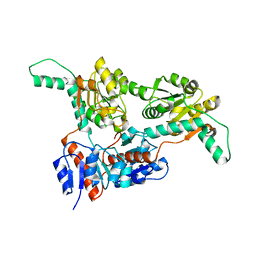 | |
6IH2
 
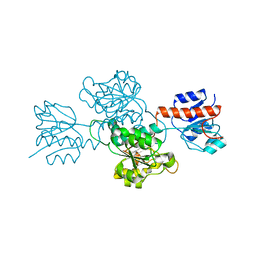 | |
6IH3
 
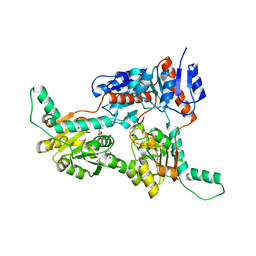 | | Crystal structure of Phosphite Dehydrogenase from Ralstonia sp. 4506 in complex with non-natural cofactor Nicotinamide Cytosine Dinucleotide | | Descriptor: | Phosphite dehydrogenase, [[(2S,3S,4R,5S)-5-(3-aminocarbonylpyridin-1-ium-1-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl] [(2S,3S,4R,5S)-5-(4-azanyl-2-oxidanylidene-pyrimidin-1-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methyl hydrogen phosphate | | Authors: | Song, X, Feng, Y, Zhao, Z, Liu, Y. | | Deposit date: | 2018-09-28 | | Release date: | 2019-03-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.942 Å) | | Cite: | Structural Insights into Phosphite Dehydrogenase Variants Favoring a Non-natural Redox Cofactor
Acs Catalysis, 9, 2019
|
|
6IH6
 
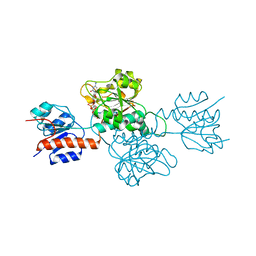 | | Phosphite Dehydrogenase mutant I151R/P176R/M207A from Ralstonia sp. 4506 in complex with non-natural cofactor Nicotinamide Cytosine dinucleotide | | Descriptor: | Phosphite dehydrogenase, [[(2S,3S,4R,5S)-5-(3-aminocarbonylpyridin-1-ium-1-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl] [(2S,3S,4R,5S)-5-(4-azanyl-2-oxidanylidene-pyrimidin-1-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methyl hydrogen phosphate | | Authors: | Song, X, Feng, Y, Liu, Y, Zhao, Z. | | Deposit date: | 2018-09-28 | | Release date: | 2019-03-13 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.491 Å) | | Cite: | Structural Insights into Phosphite Dehydrogenase Variants Favoring a Non-natural Redox Cofactor
Acs Catalysis, 9, 2019
|
|
6IH8
 
 | |
7DYS
 
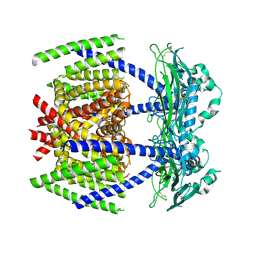 | |
8X1W
 
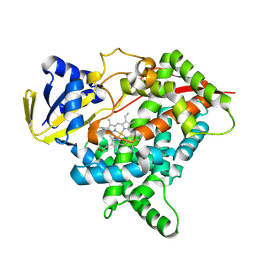 | | CYP725A4 apo structure | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, Taxadiene 5-alpha hydroxylase | | Authors: | Chang, Z, Wang, Q. | | Deposit date: | 2023-11-09 | | Release date: | 2024-08-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Unraveling the Catalytic Mechanism of Taxadiene-5alpha-hydroxylase from Crystallography and Computational Analyses.
Acs Catalysis, 14, 2024
|
|
8X3E
 
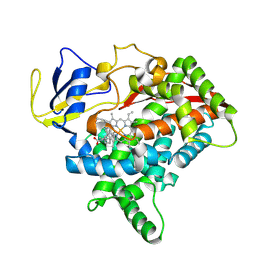 | | CYP725A4-Taxa-4,11-diene complex | | Descriptor: | (1~{R},3~{R},8~{R})-4,8,12,15,15-pentamethyltricyclo[9.3.1.0^{3,8}]pentadeca-4,11-diene, PROTOPORPHYRIN IX CONTAINING FE, Taxadiene 5-alpha hydroxylase | | Authors: | Chang, Z, Wang, Q. | | Deposit date: | 2023-11-13 | | Release date: | 2024-08-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Unraveling the Catalytic Mechanism of Taxadiene-5alpha-hydroxylase from Crystallography and Computational Analyses.
Acs Catalysis, 14, 2024
|
|
8IA1
 
 | |
8IMS
 
 | | Crystal structure of TRAF7 coiled-coil domain | | Descriptor: | E3 ubiquitin-protein ligase TRAF7 | | Authors: | Hu, R, Lin, L, Lu, Q. | | Deposit date: | 2023-03-07 | | Release date: | 2024-01-24 | | Last modified: | 2024-07-17 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | The structure of TRAF7 coiled-coil trimer provides insight into its function in zebrafish embryonic development.
J Mol Cell Biol, 16, 2024
|
|
1KO6
 
 | | Crystal Structure of C-terminal Autoproteolytic Domain of Nucleoporin Nup98 | | Descriptor: | Nuclear Pore Complex Protein Nup98 | | Authors: | Hodel, A.E, Hodel, M.R, Griffis, E.R, Hennig, K.A, Ratner, G.A, Songli, X, Powers, M.A. | | Deposit date: | 2001-12-20 | | Release date: | 2002-09-04 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The three-dimensional structure of the autoproteolytic, nuclear pore-targeting domain of the human nucleoporin Nup98.
Mol.Cell, 10, 2002
|
|
4W8D
 
 | | Crystal structure of MST3 with a pyrrolopyrimidine inhibitor (PF-06454589). | | Descriptor: | 5-(1-methyl-1H-pyrazol-4-yl)-4-(morpholin-4-yl)-7H-pyrrolo[2,3-d]pyrimidine, Serine/threonine-protein kinase 24 36 kDa subunit | | Authors: | Jasti, J, Song, X, Griffor, M, Kurumbail, R.G. | | Deposit date: | 2014-08-24 | | Release date: | 2015-03-18 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Discovery and preclinical profiling of 3-[4-(morpholin-4-yl)-7H-pyrrolo[2,3-d]pyrimidin-5-yl]benzonitrile (PF-06447475), a highly potent, selective, brain penetrant, and in vivo active LRRK2 kinase inhibitor.
J.Med.Chem., 58, 2015
|
|
8Z03
 
 | | Lactate bound to human GTP-specific succinyl-CoA synthetase | | Descriptor: | (2S)-2-HYDROXYPROPANOIC ACID, COENZYME A, MAGNESIUM ION, ... | | Authors: | Liu, R.L, Ren, X.L, Li, L.T, Zhang, Y, Huang, H, Zhao, Y.M. | | Deposit date: | 2024-04-08 | | Release date: | 2024-12-11 | | Last modified: | 2025-02-19 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Nuclear GTPSCS functions as a lactyl-CoA synthetase to promote histone lactylation and gliomagenesis.
Cell Metab., 37, 2025
|
|
8Z02
 
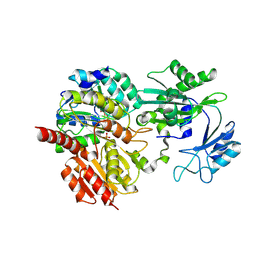 | | CoA bound to human GTP-specific succinyl-CoA synthetase | | Descriptor: | COENZYME A, PHOSPHATE ION, Succinate--CoA ligase [ADP/GDP-forming] subunit alpha, ... | | Authors: | Liu, R.L, Ren, X.L, Li, L.T, Zhang, Y, Huang, H, Zhao, Y.M. | | Deposit date: | 2024-04-08 | | Release date: | 2024-12-11 | | Last modified: | 2025-02-19 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Nuclear GTPSCS functions as a lactyl-CoA synthetase to promote histone lactylation and gliomagenesis.
Cell Metab., 37, 2025
|
|
9KNI
 
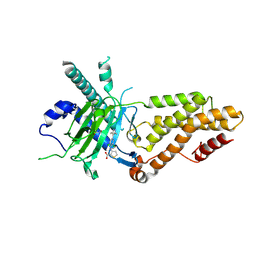 | | Structural complex of FTO bound with 12j | | Descriptor: | 2-OXOGLUTARIC ACID, 3-[[2,6-bis(chloranyl)-4-(3,5-dimethyl-1H-pyrazol-4-yl)phenyl]amino]thiophene-2-carboxylic acid, Alpha-ketoglutarate-dependent dioxygenase FTO | | Authors: | Yang, C.G, Gan, J.H. | | Deposit date: | 2024-11-19 | | Release date: | 2025-03-12 | | Method: | X-RAY DIFFRACTION (2.63 Å) | | Cite: | Development of 3-arylaminothiophenic-2-carboxylic acid derivatives as new FTO inhibitors showing potent antileukemia activities.
Eur.J.Med.Chem., 289, 2025
|
|
6KPC
 
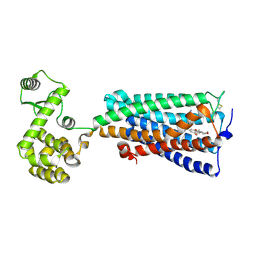 | | Crystal structure of an agonist bound GPCR | | Descriptor: | 7-[(6aR,9R,10aR)-1-Hydroxy-9-(hydroxymethyl)-6,6-dimethyl-6a,7,8,9,10,10a-hexahydro-6H-benzo[c]chromen-3-yl]- 7-methyloctanenitrile, Cannabinoid receptor 2,Endolysin | | Authors: | Li, X.T, Hua, T, Wu, L.J, Makriyannis, A, Wu, M, Liu, Z.J. | | Deposit date: | 2019-08-15 | | Release date: | 2020-02-12 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Activation and Signaling Mechanism Revealed by Cannabinoid Receptor-GiComplex Structures.
Cell, 180, 2020
|
|
6KPF
 
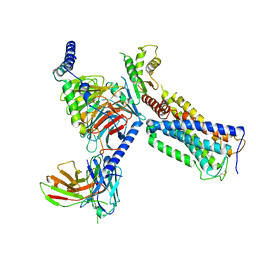 | | Cryo-EM structure of a class A GPCR with G protein complex | | Descriptor: | 7-[(6aR,9R,10aR)-1-Hydroxy-9-(hydroxymethyl)-6,6-dimethyl-6a,7,8,9,10,10a-hexahydro-6H-benzo[c]chromen-3-yl]- 7-methyloctanenitrile, Cannabinoid receptor 2, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Li, X.T, Hua, T, Wu, L.J, Makriyannis, A, Shen, L, Wang, Y.X, Liu, Z.J. | | Deposit date: | 2019-08-15 | | Release date: | 2020-02-12 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Activation and Signaling Mechanism Revealed by Cannabinoid Receptor-GiComplex Structures.
Cell, 180, 2020
|
|
