7R20
 
 | | Anti-Arc nanobody E5 | | Descriptor: | Anti-Arc nanobody E5, GLYCEROL, SULFATE ION | | Authors: | Markusson, S, Kursula, P. | | Deposit date: | 2022-02-03 | | Release date: | 2022-06-01 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | High-affinity anti-Arc nanobodies provide tools for structural and functional studies.
Plos One, 17, 2022
|
|
7R1Z
 
 | |
7R23
 
 | |
6H42
 
 | | crystal structure of the human TGT catalytic subunit QTRT1 | | Descriptor: | BROMIDE ION, CHLORIDE ION, GLUTAMIC ACID, ... | | Authors: | Johannsson, S, Neumann, P, Ficner, R. | | Deposit date: | 2018-07-20 | | Release date: | 2018-09-05 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Crystal Structure of the Human tRNA Guanine Transglycosylase Catalytic Subunit QTRT1.
Biomolecules, 8, 2018
|
|
6H45
 
 | | crystal structure of the human TGT catalytic subunit QTRT1 in complex with queuine | | Descriptor: | 2-amino-5-({[(1S,4S,5R)-4,5-dihydroxycyclopent-2-en-1-yl]amino}methyl)-3,7-dihydro-4H-pyrrolo[2,3-d]pyrimidin-4-one, BROMIDE ION, CHLORIDE ION, ... | | Authors: | Johannsson, S, Neumann, P, Ficner, R. | | Deposit date: | 2018-07-20 | | Release date: | 2018-09-05 | | Last modified: | 2023-03-15 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of the Human tRNA Guanine Transglycosylase Catalytic Subunit QTRT1.
Biomolecules, 8, 2018
|
|
4AYI
 
 | | Structure of a complex between CCPs 6 and 7 of Human Complement Factor H and Neisseria meningitidis FHbp Variant 3 Wild type | | Descriptor: | 1,2-ETHANEDIOL, COMPLEMENT FACTOR H, LIPOPROTEIN GNA1870 CCOMPND 7 | | Authors: | Johnson, S, Tan, L, van der Veen, S, Caesar, J, Goicoechea De Jorge, E, Everett, R.J, Bai, X, Exley, R.M, Ward, P.N, Ruivo, N, Trivedi, K, Cumber, E, Jones, R, Newham, L, Staunton, D, Borrow, R, Pickering, M, Lea, S.M, Tang, C.M. | | Deposit date: | 2012-06-21 | | Release date: | 2012-11-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Design and Evaluation of Meningococcal Vaccines Through Structure-Based Modification of Host and Pathogen Molecules.
Plos Pathog., 8, 2012
|
|
4AYN
 
 | | Structure of the C-terminal barrel of Neisseria meningitidis FHbp Variant 2 | | Descriptor: | FACTOR H-BINDING PROTEIN, SULFATE ION | | Authors: | Johnson, S, Tan, L, van der Veen, S, Caesar, J, Goicoechea De Jorge, E, Everett, R.J, Bai, X, Exley, R.M, Ward, P.N, Ruivo, N, Trivedi, K, Cumber, E, Jones, R, Newham, L, Staunton, D, Borrow, R, Pickering, M, Lea, S.M, Tang, C.M. | | Deposit date: | 2012-06-21 | | Release date: | 2012-11-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Design and Evaluation of Meningococcal Vaccines Through Structure-Based Modification of Host and Pathogen Molecules.
Plos Pathog., 8, 2012
|
|
4AYD
 
 | | Structure of a complex between CCPs 6 and 7 of Human Complement Factor H and Neisseria meningitidis FHbp Variant 1 R106A mutant | | Descriptor: | 1,2-ETHANEDIOL, COMPLEMENT FACTOR H, FACTOR H BINDING PROTEIN | | Authors: | Johnson, S, Tan, L, van der Veen, S, Caesar, J, Goicoechea De Jorge, E, Everett, R.J, Bai, X, Exley, R.M, Ward, P.N, Ruivo, N, Trivedi, K, Cumber, E, Jones, R, Newham, L, Staunton, D, Borrow, R, Pickering, M, Lea, S.M, Tang, C.M. | | Deposit date: | 2012-06-20 | | Release date: | 2012-11-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Design and Evaluation of Meningococcal Vaccines Through Structure-Based Modification of Host and Pathogen Molecules
Plos Pathog., 8, 2012
|
|
4AYE
 
 | | Structure of a complex between CCPs 6 and 7 of Human Complement Factor H and Neisseria meningitidis FHbp Variant 1 E283AE304A mutant | | Descriptor: | 1,2-ETHANEDIOL, COMPLEMENT FACTOR H, FACTOR H BINDING PROTEIN | | Authors: | Johnson, S, Tan, L, van der Veen, S, Caesar, J, Goicoechea De Jorge, E, Everett, R.J, Bai, X, Exley, R.M, Ward, P.N, Ruivo, N, Trivedi, K, Cumber, E, Jones, R, Newham, L, Staunton, D, Borrow, R, Pickering, M, Lea, S.M, Tang, C.M. | | Deposit date: | 2012-06-20 | | Release date: | 2012-11-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Design and Evaluation of Meningococcal Vaccines Through Structure-Based Modification of Host and Pathogen Molecules.
Plos Pathog., 8, 2012
|
|
4AYM
 
 | | Structure of a complex between CCPs 6 and 7 of Human Complement Factor H and Neisseria meningitidis FHbp Variant 3 P106A mutant | | Descriptor: | COMPLEMENT FACTOR H, FACTOR H BINDING PROTEIN | | Authors: | Johnson, S, Tan, L, van der Veen, S, Caesar, J, Goicoechea De Jorge, E, Everett, R.J, Bai, X, Exley, R.M, Ward, P.N, Ruivo, N, Trivedi, K, Cumber, E, Jones, R, Newham, L, Staunton, D, Borrow, R, Pickering, M, Lea, S.M, Tang, C.M. | | Deposit date: | 2012-06-21 | | Release date: | 2012-11-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Design and Evaluation of Meningococcal Vaccines Through Structure-Based Modification of Host and Pathogen Molecules.
Plos Pathog., 8, 2012
|
|
1OTA
 
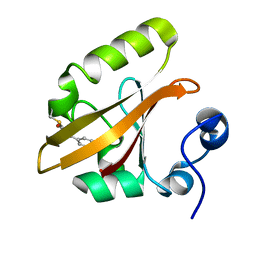 | | E46Q MUTANT OF PHOTOACTIVE YELLOW PROTEIN, P63 AT 295K | | Descriptor: | 4'-HYDROXYCINNAMIC ACID, Photoactive yellow protein | | Authors: | Anderson, S, Crosson, S, Moffat, K. | | Deposit date: | 2003-03-21 | | Release date: | 2004-05-11 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Short hydrogen bonds in photoactive yellow protein.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
4BIK
 
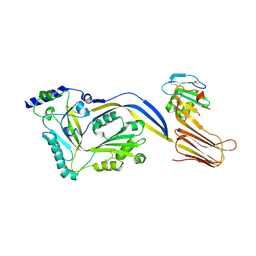 | | Structure of a disulfide locked mutant of Intermedilysin with human CD59 | | Descriptor: | CD59 GLYCOPROTEIN, INTERMEDILYSIN | | Authors: | Johnson, S, Brooks, N.J, Smith, R.A.G, Lea, S.M, Bubeck, D. | | Deposit date: | 2013-04-10 | | Release date: | 2013-05-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.494 Å) | | Cite: | Structural Basis for Recognition of the Pore- Forming Toxin Intermedilysin by Human Complement Receptor Cd59
Cell Rep., 3, 2013
|
|
2WGU
 
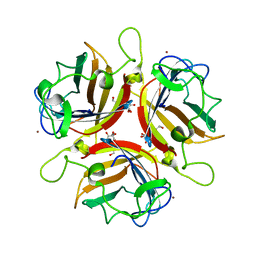 | | Structure of human adenovirus serotype 37 fibre head in complex with a sialic acid derivative, O-Methyl 5-N- methoxycarbonyl -3,5-dideoxy- D-glycero-a-D-galacto-2-nonulopyranosylonic acid | | Descriptor: | 3,5-dideoxy-5-[(methoxycarbonyl)amino]-D-glycero-alpha-D-galacto-non-2-ulopyranosonic acid, FIBER PROTEIN, ZINC ION | | Authors: | Johansson, S, Nilsson, E, Qian, W, Guilligay, D, Crepin, T, Cusack, S, Arnberg, N, Elofsson, M. | | Deposit date: | 2009-04-27 | | Release date: | 2009-11-24 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Design, Synthesis, and Evaluation of N-Acyl Modified Sialic Acids as Inhibitors of Adenoviruses Causing Epidemic Keratoconjunctivitis.
J.Med.Chem., 52, 2009
|
|
2WGT
 
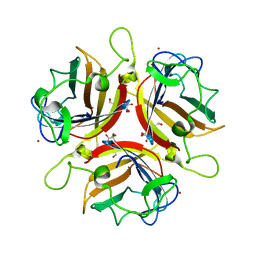 | | Structure of human adenovirus serotype 37 fibre head in complex with a sialic acid derivative, O-Methyl 5-N-propaonyl-3,5-dideoxy-D- glycero-a-D-galacto-2-nonulopyranosylonic acid | | Descriptor: | 3,5-dideoxy-5-(propanoylamino)-D-glycero-alpha-D-galacto-non-2-ulopyranosonic acid, FIBER PROTEIN, ZINC ION | | Authors: | Johansson, S, Nilsson, E, Qian, W, Guilligay, D, Crepin, T, Cusack, S, Arnberg, N, Elofsson, M. | | Deposit date: | 2009-04-27 | | Release date: | 2009-11-24 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Design, Synthesis, and Evaluation of N-Acyl Modified Sialic Acids as Inhibitors of Adenoviruses Causing Epidemic Keratoconjunctivitis.
J.Med.Chem., 52, 2009
|
|
7BK0
 
 | |
7BJ2
 
 | |
7BGL
 
 | | Salmonella LP ring 26 mer refined in C26 map | | Descriptor: | (2~{R},4~{R},5~{R},6~{R})-6-[(1~{R})-1,2-bis(oxidanyl)ethyl]-4,5-bis(oxidanyl)oxane-2-carboxylic acid, Flagellar L-ring protein, Flagellar P-ring protein, ... | | Authors: | Johnson, S, Furlong, E, Lea, S.M. | | Deposit date: | 2021-01-07 | | Release date: | 2021-05-05 | | Last modified: | 2021-07-14 | | Method: | ELECTRON MICROSCOPY (2.2 Å) | | Cite: | Molecular structure of the intact bacterial flagellar basal body.
Nat Microbiol, 6, 2021
|
|
7BIN
 
 | | Salmonella export gate and rod refined in focussed C1 map | | Descriptor: | Flagellar basal body rod protein FlgB, Flagellar basal-body rod protein FlgC, Flagellar basal-body rod protein FlgF, ... | | Authors: | Johnson, S, Furlong, E, Lea, S.M. | | Deposit date: | 2021-01-12 | | Release date: | 2021-05-05 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Molecular structure of the intact bacterial flagellar basal body.
Nat Microbiol, 6, 2021
|
|
7BHQ
 
 | |
6TAS
 
 | | Structure of the two-fold capsomer of the dArc1 capsid | | Descriptor: | Activity-regulated cytoskeleton associated protein 1, ZINC ION | | Authors: | Erlendsson, S, Morado, D.R, Shepherd, J.D, Briggs, J.A.G. | | Deposit date: | 2019-10-30 | | Release date: | 2020-01-01 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (2.75 Å) | | Cite: | Structures of virus-like capsids formed by the Drosophila neuronal Arc proteins.
Nat.Neurosci., 23, 2020
|
|
6TAQ
 
 | |
6TAU
 
 | |
6TAR
 
 | |
6TAT
 
 | |
6TAP
 
 | | Structure of the dArc1 capsid | | Descriptor: | Activity-regulated cytoskeleton associated protein 1, ZINC ION | | Authors: | Erlendsson, S, Morado, D.R, Shepherd, J.D, Briggs, J.A.G. | | Deposit date: | 2019-10-30 | | Release date: | 2020-01-01 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structures of virus-like capsids formed by the Drosophila neuronal Arc proteins.
Nat.Neurosci., 23, 2020
|
|
