3MER
 
 | | Crystal Structure of the methyltransferase Slr1183 from Synechocystis sp. PCC 6803, Northeast Structural Genomics Consortium Target SgR145 | | Descriptor: | Slr1183 protein | | Authors: | Seetharaman, J, Chen, Y, Forouhar, F, Rossi, P, Sahdev, S, Xiao, R, Ciccosanti, C, Foote, E.L, Belote, R.L, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2010-03-31 | | Release date: | 2010-04-14 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Northeast Structural Genomics Consortium Target SgR145
To be Published
|
|
5JTN
 
 | | The structure of chaperone SecB in complex with unstructured proPhoA binding site c | | Descriptor: | Alkaline phosphatase, Protein-export protein SecB | | Authors: | Huang, C, Saio, T, Rossi, P, Kalodimos, C.G. | | Deposit date: | 2016-05-09 | | Release date: | 2016-08-24 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural basis for the antifolding activity of a molecular chaperone.
Nature, 537, 2016
|
|
5JTL
 
 | | The structure of chaperone SecB in complex with unstructured proPhoA | | Descriptor: | Alkaline phosphatase, Protein-export protein SecB | | Authors: | Huang, C, Saio, T, Rossi, P, Kalodimos, C.G. | | Deposit date: | 2016-05-09 | | Release date: | 2016-08-24 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural basis for the antifolding activity of a molecular chaperone.
Nature, 537, 2016
|
|
5JTM
 
 | | The structure of chaperone SecB in complex with unstructured PhoA binding site a | | Descriptor: | Alkaline phosphatase, Protein-export protein SecB | | Authors: | Huang, C, Saio, T, Rossi, P, Kalodimos, C.G. | | Deposit date: | 2016-05-09 | | Release date: | 2016-08-24 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural basis for the antifolding activity of a molecular chaperone.
Nature, 537, 2016
|
|
5JTQ
 
 | | The structure of chaperone SecB in complex with unstructured MBP binding site d | | Descriptor: | Maltose-binding periplasmic protein, Protein-export protein SecB | | Authors: | Huang, C, Saio, T, Rossi, P, Kalodimos, C.G. | | Deposit date: | 2016-05-09 | | Release date: | 2016-08-24 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural basis for the antifolding activity of a molecular chaperone.
Nature, 537, 2016
|
|
5JTO
 
 | | The structure of chaperone SecB in complex with unstructured proPhoA binding site d | | Descriptor: | Alkaline phosphatase, Protein-export protein SecB | | Authors: | Huang, C, Saio, T, Rossi, P, Kalodimos, C.G. | | Deposit date: | 2016-05-09 | | Release date: | 2016-08-24 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural basis for the antifolding activity of a molecular chaperone.
Nature, 537, 2016
|
|
5JTP
 
 | | The structure of chaperone SecB in complex with unstructured proPhoA binding site e | | Descriptor: | Alkaline phosphatase, Protein-export protein SecB | | Authors: | Huang, C, Saio, T, Rossi, P, Kalodimos, C.G. | | Deposit date: | 2016-05-09 | | Release date: | 2016-08-24 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural basis for the antifolding activity of a molecular chaperone.
Nature, 537, 2016
|
|
5JTR
 
 | | The structure of chaperone SecB in complex with unstructured MBP binding site e | | Descriptor: | Maltose-binding periplasmic protein, Protein-export protein SecB | | Authors: | Huang, C, Saio, T, Rossi, P, Kalodimos, C.G. | | Deposit date: | 2016-05-09 | | Release date: | 2016-08-24 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural basis for the antifolding activity of a molecular chaperone.
Nature, 537, 2016
|
|
5KP0
 
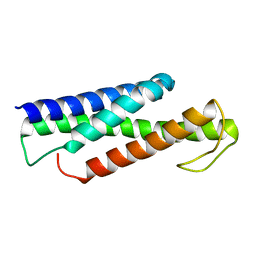 | | Recognition and targeting mechanisms by chaperones in flagella assembly and operation | | Descriptor: | Flagellar protein FliT,Flagellum-specific ATP synthase | | Authors: | Khanra, N.K, Rossi, P, Economou, A, Kalodimos, C.G. | | Deposit date: | 2016-07-01 | | Release date: | 2016-08-17 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Recognition and targeting mechanisms by chaperones in flagellum assembly and operation.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5KS6
 
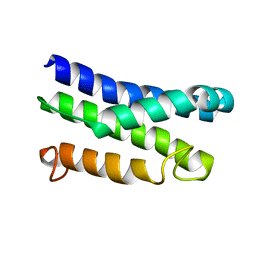 | |
3FIF
 
 | | Crystal structure of the ygdR protein from E.coli. Northeast Structural Genomics target ER382A. | | Descriptor: | Uncharacterized ligand, Uncharacterized lipoprotein ygdR | | Authors: | Kuzin, A.P, Su, M, Seetharaman, J, Rossi, P, Chen, C.X, Jiang, M, Cunningham, K, Ma, L, Xiao, R, Liu, J.C, Baran, M, Swapna, G.V.T, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-12-11 | | Release date: | 2009-01-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of the ygdR protein from E.coli. Northeast Structural Genomics target ER382A.
To be Published
|
|
1MMI
 
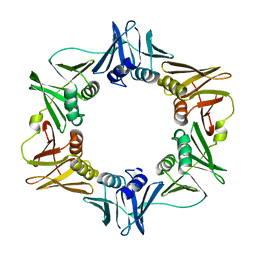 | | E. COLI DNA POLYMERASE BETA SUBUNIT | | Descriptor: | DNA polymerase III, beta chain | | Authors: | Oakley, A.J, Prosselkov, P, Wijffels, G, Beck, J.L, Wilce, M.C.J, Dixon, N.E. | | Deposit date: | 2002-09-04 | | Release date: | 2003-09-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.848 Å) | | Cite: | Flexibility revealed by the 1.85 A crystal structure of the beta sliding-clamp subunit of Escherichia coli DNA polymerase III.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
3IPF
 
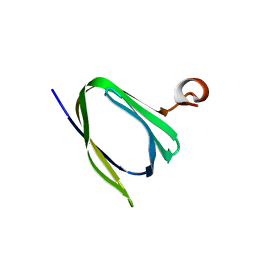 | | Crystal structure of the Q251Q8_DESHY protein from Desulfitobacterium hafniense. Northeast Structural Genomics Consortium Target DhR8c. | | Descriptor: | uncharacterized protein | | Authors: | Vorobiev, S.M, Chen, Y, Seetharaman, J, Janjua, H, Xiao, R, Ciccosanti, C, Wang, H, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Andre, I, Rossi, P, Kennedy, M, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-08-17 | | Release date: | 2009-09-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.988 Å) | | Cite: | Crystal structure of the Q251Q8_DESHY protein from Desulfitobacterium hafniense.
To be Published
|
|
6XR6
 
 | |
6XR7
 
 | |
6XRG
 
 | |
3LMO
 
 | | Crystal Structure of specialized acyl carrier protein (RPA2022) from Rhodopseudomonas palustris, Northeast Structural Genomics Consortium Target RpR324 | | Descriptor: | Specialized acyl carrier protein | | Authors: | Forouhar, F, Rossi, P, Lew, S, Seetharaman, J, Mao, M, Xiao, R, Ciccosanti, C, Wang, H, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2010-01-31 | | Release date: | 2010-02-16 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of a specialized acyl carrier protein essential for lipid A biosynthesis with very long-chain fatty acids in open and closed conformations.
Biochemistry, 51, 2012
|
|
7N00
 
 | | Anaplastic lymphoma kinase (ALK) extracellular fragment of ligand binding region 648-1025 in complex with AUG-alpha | | Descriptor: | ALK and LTK ligand 2, ALK tyrosine kinase receptor | | Authors: | Reshetnyak, A.V, Myasnikov, A.G, Rossi, P, Miller, D.J, Kalodimos, C.G. | | Deposit date: | 2021-05-24 | | Release date: | 2021-11-24 | | Last modified: | 2021-12-15 | | Method: | ELECTRON MICROSCOPY (2.27 Å) | | Cite: | Mechanism for the activation of the anaplastic lymphoma kinase receptor.
Nature, 600, 2021
|
|
7MZY
 
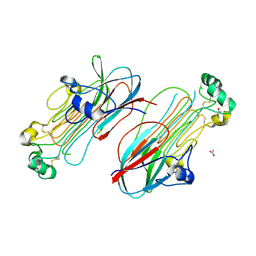 | | Anaplastic lymphoma kinase (ALK) extracellular fragment of ligand binding region 673-986 | | Descriptor: | ACETATE ION, ALK tyrosine kinase receptor | | Authors: | Reshetnyak, A.V, Sowaileh, M, Miller, D.J, Rossi, P, Myasnikov, A.G, Kalodimos, C.G. | | Deposit date: | 2021-05-24 | | Release date: | 2021-11-24 | | Last modified: | 2021-12-15 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Mechanism for the activation of the anaplastic lymphoma kinase receptor.
Nature, 600, 2021
|
|
1K2B
 
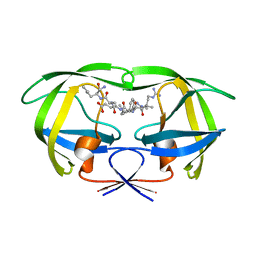 | | Combining Mutations in HIV-1 Protease to Understand Mechanisms of Resistance | | Descriptor: | N-[(2R)-2-({N~5~-[amino(iminio)methyl]-L-ornithyl-L-valyl}amino)-4-methylpentyl]-L-phenylalanyl-L-alpha-glutamyl-L-alanyl-L-norleucinamide, PROTEASE RETROPEPSIN | | Authors: | Mahalingam, B, Boross, P, Wang, Y.-F, Louis, J.M, Fischer, C, Tozser, J, W Harrison, R, Weber, I.T. | | Deposit date: | 2001-09-26 | | Release date: | 2002-07-10 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Combining mutations in HIV-1 protease to understand mechanisms of resistance.
Proteins, 48, 2002
|
|
6AMW
 
 | |
6AMV
 
 | |
1K1T
 
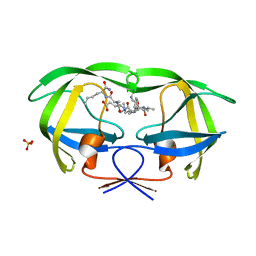 | | Combining Mutations in HIV-1 Protease to Understand Mechanisms of Resistance | | Descriptor: | N-[(2R)-2-({N~5~-[amino(iminio)methyl]-L-ornithyl-L-valyl}amino)-4-methylpentyl]-L-phenylalanyl-L-alpha-glutamyl-L-alanyl-L-norleucinamide, PROTEASE RETROPEPSIN, SULFATE ION | | Authors: | Mahalingam, B, Boross, P, Wang, Y.-F, Louis, J.M, Fischer, C, Tozser, J, Harrison, R.W, Weber, I.T. | | Deposit date: | 2001-09-25 | | Release date: | 2002-07-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Combining mutations in HIV-1 protease to understand mechanisms of resistance.
Proteins, 48, 2002
|
|
1K2C
 
 | | Combining Mutations in HIV-1 Protease to Understand Mechanisms of Resistance | | Descriptor: | N-[(2R)-2-({N~5~-[amino(iminio)methyl]-L-ornithyl-L-valyl}amino)-4-methylpentyl]-L-phenylalanyl-L-alpha-glutamyl-L-alanyl-L-norleucinamide, PROTEASE RETROPEPSIN | | Authors: | Mahalingam, B, Boross, P, Wang, Y.-F, Louis, J.M, Fischer, C, Tozser, J, W Harrison, R, Weber, I.T. | | Deposit date: | 2001-09-26 | | Release date: | 2002-07-10 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Combining mutations in HIV-1 protease to understand mechanisms of resistance.
Proteins, 48, 2002
|
|
3QSB
 
 | | Structure of E. coli polIIIbeta with (Z)-5-(1-((4'-Fluorobiphenyl-4-yl)methoxyimino)butyl)-2,2-dimethyl-4,6-dioxocyclohexanecarbonitrile | | Descriptor: | (1R,5R)-5-{(1Z)-N-[(4'-fluorobiphenyl-4-yl)methoxy]butanimidoyl}-2,2-dimethyl-4,6-dioxocyclohexanecarbonitrile, DNA polymerase III subunit beta | | Authors: | Wijffels, G, Johnson, W.M, Oakley, A.J, Turner, K, Epa, V.C, Briscoe, S.J, Polley, M, Liepa, A.J, Hofmann, A, Buchardt, J, Christensen, C, Prosselkov, P, Dalrymple, B.P, Alewood, P.F, Jennings, P.A, Dixon, N.E, Winkler, D.A. | | Deposit date: | 2011-02-20 | | Release date: | 2011-06-08 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Binding inhibitors of the bacterial sliding clamp by design
J.Med.Chem., 54, 2011
|
|
