5I8Y
 
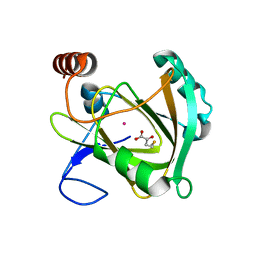 | | Structure of Mouse Acireductone Dioxygenase bound to Co2+ and 2-keto-4-(methylthio)-butyric acid | | Descriptor: | 1,2-dihydroxy-3-keto-5-methylthiopentene dioxygenase, 4-(METHYLSULFANYL)-2-OXOBUTANOIC ACID, COBALT (II) ION | | Authors: | Deshpande, A.R, Wagenpfeil, K, Pochapsky, T.C, Petsko, G.A, Ringe, D. | | Deposit date: | 2016-02-19 | | Release date: | 2016-03-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.942 Å) | | Cite: | Metal-Dependent Function of a Mammalian Acireductone Dioxygenase.
Biochemistry, 55, 2016
|
|
5I93
 
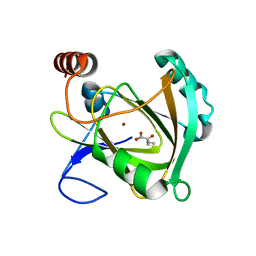 | | Structure of Mouse Acireductone dioxygenase with Ni2+ and 2-ketopentanoic acid in the active site | | Descriptor: | 1,2-dihydroxy-3-keto-5-methylthiopentene dioxygenase, 2-oxopentanoic acid, NICKEL (II) ION | | Authors: | Deshpande, A.R, Wagenpfeil, K, Pochapsky, T.C, Petsko, G.A, Ringe, D. | | Deposit date: | 2016-02-19 | | Release date: | 2016-03-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.236 Å) | | Cite: | Metal-Dependent Function of a Mammalian Acireductone Dioxygenase.
Biochemistry, 55, 2016
|
|
5I91
 
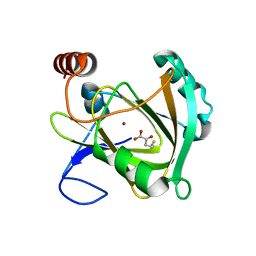 | | Structure of Mouse Acirecutone dioxygenase with to Ni2+ and 2-keto-4-(methylthio)-butyric acid in the active site | | Descriptor: | 1,2-dihydroxy-3-keto-5-methylthiopentene dioxygenase, 4-(METHYLSULFANYL)-2-OXOBUTANOIC ACID, NICKEL (II) ION | | Authors: | Deshpande, A.R, Robinson, H, Wagenpfeil, K, Pochapsky, T.C, Petsko, G.A, Ringe, D. | | Deposit date: | 2016-02-19 | | Release date: | 2016-03-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Metal-Dependent Function of a Mammalian Acireductone Dioxygenase.
Biochemistry, 55, 2016
|
|
5I8S
 
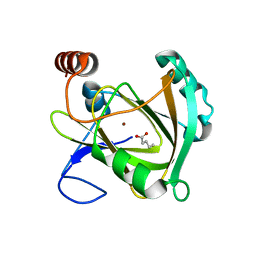 | | Structure of Mouse Acireductone dioxygenase with Ni2+ ion and pentanoic acid in the active site | | Descriptor: | 1,2-dihydroxy-3-keto-5-methylthiopentene dioxygenase, NICKEL (II) ION, PENTANOIC ACID | | Authors: | Deshpande, A.R, Wagenpfeil, K, Pochapsky, T.C, Petsko, G.A, Ringe, D. | | Deposit date: | 2016-02-19 | | Release date: | 2016-03-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Metal-Dependent Function of a Mammalian Acireductone Dioxygenase.
Biochemistry, 55, 2016
|
|
6C2Q
 
 | | Crystal Structures of Cystathionine beta-Synthase from Saccharomyces cerevisiae: the Structure of the PLP-L-Serine Intermediate | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Kreinbring, C.A, Tu, Y, Liu, D, Petsko, G.A, Ringe, D. | | Deposit date: | 2018-01-08 | | Release date: | 2018-04-25 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Crystal Structures of Cystathionine beta-Synthase from Saccharomyces cerevisiae: One Enzymatic Step at a Time.
Biochemistry, 57, 2018
|
|
6C2H
 
 | | Crystal Structures of Cystathionine beta-Synthase from Saccharomyces cerevisiae: the Structure of the Catalytic Core | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, CALCIUM ION, ... | | Authors: | Kreinbring, C.A, Tu, Y, Liu, D, Petsko, G.A, Ringe, D. | | Deposit date: | 2018-01-08 | | Release date: | 2018-04-25 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Crystal Structures of Cystathionine beta-Synthase from Saccharomyces cerevisiae: One Enzymatic Step at a Time.
Biochemistry, 57, 2018
|
|
6C4P
 
 | | Crystal Structures of Cystathionine beta-Synthase from Saccharomyces cerevisiae: the Structure of the PMP Complex | | Descriptor: | 1,2-ETHANEDIOL, 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, CALCIUM ION, ... | | Authors: | Kreinbring, C.A, Tu, Y, Liu, D, Berkowitz, D.B, Petsko, G.A, Ringe, D. | | Deposit date: | 2018-01-12 | | Release date: | 2018-04-25 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structures of Cystathionine beta-Synthase from Saccharomyces cerevisiae: One Enzymatic Step at a Time.
Biochemistry, 57, 2018
|
|
1A0G
 
 | | L201A MUTANT OF D-AMINO ACID AMINOTRANSFERASE COMPLEXED WITH PYRIDOXAMINE-5'-PHOSPHATE | | Descriptor: | 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, D-AMINO ACID AMINOTRANSFERASE | | Authors: | Sugio, S, Kashima, A, Kishimoto, K, Peisach, D, Petsko, G.A, Ringe, D, Yoshimura, T, Esaki, N. | | Deposit date: | 1997-11-30 | | Release date: | 1998-06-03 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of L201A mutant of D-amino acid aminotransferase at 2.0 A resolution: implication of the structural role of Leu201 in transamination.
Protein Eng., 11, 1998
|
|
3QYH
 
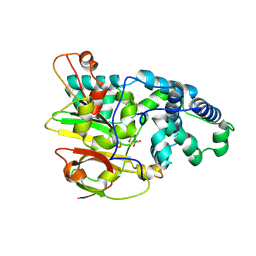 | | Crystal Structure of Co-type Nitrile Hydratase beta-H71L from Pseudomonas putida. | | Descriptor: | COBALT (III) ION, Co-type Nitrile Hydratase alpha subunit, Co-type Nitrile Hydratase beta subunit | | Authors: | Brodkin, H.R, Novak, W.R.P, Ringe, D, Petsko, G.A. | | Deposit date: | 2011-03-03 | | Release date: | 2011-03-23 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Evidence of the Participation of Remote Residues in the Catalytic Activity of Co-Type Nitrile Hydratase from Pseudomonas putida.
Biochemistry, 50, 2011
|
|
3F6E
 
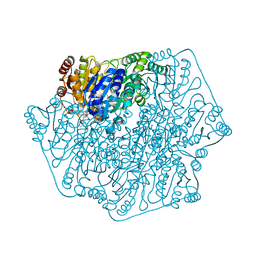 | | Crystal structure of benzoylformate decarboxylase in complex with the pyridyl inhibitor 3-PKB | | Descriptor: | 3-[(4-amino-2-methylpyrimidin-5-yl)methyl]-5-(2-{[(S)-hydroxy(phosphonooxy)phosphoryl]oxy}ethyl)-2-[(1S,2E)-1-hydroxy-3-pyridin-3-ylprop-2-en-1-yl]-4-methyl-1,3-thiazol-3-ium, Benzoylformate decarboxylase, MAGNESIUM ION | | Authors: | Brandt, G.S, McLeish, M.J, Kenyon, G.L, Petsko, G.A, Ringe, D, Jordan, F. | | Deposit date: | 2008-11-05 | | Release date: | 2008-12-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Detection and time course of formation of major thiamin diphosphate-bound covalent intermediates derived from a chromophoric substrate analogue on benzoylformate decarboxylase.
Biochemistry, 48, 2009
|
|
3QZ9
 
 | | Crystal structure of Co-type nitrile hydratase beta-Y215F from Pseudomonas putida. | | Descriptor: | COBALT (III) ION, Co-type Nitrile Hydratase alpha subunit, Co-type Nitrile Hydratase beta subunit, ... | | Authors: | Brodkin, H.R, Novak, W.R.P, Ringe, D, Petsko, G.A. | | Deposit date: | 2011-03-04 | | Release date: | 2011-03-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Evidence of the Participation of Remote Residues in the Catalytic Activity of Co-Type Nitrile Hydratase from Pseudomonas putida.
Biochemistry, 50, 2011
|
|
3QYG
 
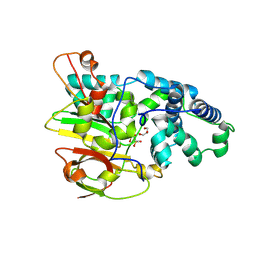 | | Crystal Structure of Co-type Nitrile Hydratase beta-E56Q from Pseudomonas putida. | | Descriptor: | COBALT (III) ION, Co-type Nitrile Hydratase alpha subunit, Co-type Nitrile Hydratase beta subunit, ... | | Authors: | Brodkin, H.R, Novak, W.R.P, Ringe, D, Petsko, G.A. | | Deposit date: | 2011-03-03 | | Release date: | 2011-03-23 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Evidence of the Participation of Remote Residues in the Catalytic Activity of Co-Type Nitrile Hydratase from Pseudomonas putida.
Biochemistry, 50, 2011
|
|
3QZ5
 
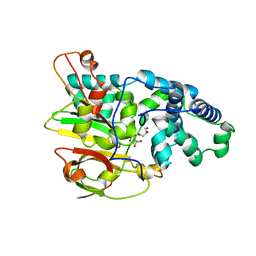 | | Crystal Structure of Co-type Nitrile Hydratase alpha-E168Q from Pseudomonas putida. | | Descriptor: | COBALT (III) ION, Co-type Nitrile Hydratase alpha subunit, Co-type Nitrile Hydratase beta subunit, ... | | Authors: | Brodkin, H.R, Novak, W.R.P, Ringe, D, Petsko, G.A. | | Deposit date: | 2011-03-04 | | Release date: | 2011-03-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Evidence of the Participation of Remote Residues in the Catalytic Activity of Co-Type Nitrile Hydratase from Pseudomonas putida.
Biochemistry, 50, 2011
|
|
3RHN
 
 | | HISTIDINE TRIAD NUCLEOTIDE-BINDING PROTEIN (HINT) FROM RABBIT COMPLEXED WITH GMP | | Descriptor: | GUANOSINE-5'-MONOPHOSPHATE, HISTIDINE TRIAD NUCLEOTIDE-BINDING PROTEIN | | Authors: | Brenner, C, Garrison, P, Gilmour, J, Peisach, D, Ringe, D, Petsko, G.A, Lowenstein, J.M. | | Deposit date: | 1997-02-11 | | Release date: | 1997-06-16 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures of HINT demonstrate that histidine triad proteins are GalT-related nucleotide-binding proteins.
Nat.Struct.Biol., 4, 1997
|
|
3F6B
 
 | | Crystal structure of benzoylformate decarboxylase in complex with the pyridyl inhibitor PAA | | Descriptor: | 3-[(4-amino-2-methylpyrimidin-5-yl)methyl]-5-(2-{[(S)-hydroxy(phosphonooxy)phosphoryl]oxy}ethyl)-2-[(1S,2E)-1-hydroxy-3-pyridin-3-ylprop-2-en-1-yl]-4-methyl-1,3-thiazol-3-ium, Benzoylformate decarboxylase, MAGNESIUM ION | | Authors: | Brandt, G.S, McLeish, M.J, Kenyon, G.L, Petsko, G.A, Ringe, D, Jordan, F. | | Deposit date: | 2008-11-05 | | Release date: | 2008-12-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Detection and time course of formation of major thiamin diphosphate-bound covalent intermediates derived from a chromophoric substrate analogue on benzoylformate decarboxylase.
Biochemistry, 48, 2009
|
|
1BFD
 
 | | BENZOYLFORMATE DECARBOXYLASE FROM PSEUDOMONAS PUTIDA | | Descriptor: | BENZOYLFORMATE DECARBOXYLASE, CALCIUM ION, MAGNESIUM ION, ... | | Authors: | Hasson, M.S, Muscate, A, Mcleish, M.J, Polovnikova, L.S, Gerlt, J.A, Kenyon, G.L, Petsko, G.A, Ringe, D. | | Deposit date: | 1998-04-30 | | Release date: | 1998-06-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The crystal structure of benzoylformate decarboxylase at 1.6 A resolution: diversity of catalytic residues in thiamin diphosphate-dependent enzymes.
Biochemistry, 37, 1998
|
|
3FSJ
 
 | | Crystal structure of benzoylformate decarboxylase in complex with the inhibitor MBP | | Descriptor: | 3-[(4-amino-2-methylpyrimidin-5-yl)methyl]-2-{(S)-hydroxy[(R)-hydroxy(methoxy)phosphoryl]phenylmethyl}-5-(2-{[(R)-hydroxy(phosphonooxy)phosphoryl]oxy}ethyl)-4-methyl-1,3-thiazol-3-ium, Benzoylformate decarboxylase, CALCIUM ION | | Authors: | Brandt, G.S, Kenyon, G.L, McLeish, M.J, Jordan, F, Petsko, G.A, Ringe, D. | | Deposit date: | 2009-01-09 | | Release date: | 2009-01-27 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | Snapshot of a reaction intermediate: analysis of benzoylformate decarboxylase in complex with a benzoylphosphonate inhibitor.
Biochemistry, 48, 2009
|
|
3QXE
 
 | | Crystal Structure of Co-type Nitrile Hydratase from Pseudomonas putida. | | Descriptor: | COBALT (III) ION, Co-type Nitrile Hydratase alpha subunit, Co-type Nitrile Hydratase beta subunit, ... | | Authors: | Brodkin, H.R, Novak, W.R.P, Ringe, D, Petsko, G.A. | | Deposit date: | 2011-03-01 | | Release date: | 2011-03-23 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.104 Å) | | Cite: | Evidence of the Participation of Remote Residues in the Catalytic Activity of Co-Type Nitrile Hydratase from Pseudomonas putida.
Biochemistry, 50, 2011
|
|
1BMA
 
 | | BENZYL METHYL AMINIMIDE INHIBITOR COMPLEXED TO PORCINE PANCREATIC ELASTASE | | Descriptor: | (1R)-1-benzyl-1-methyl-1-(2-{[4-(1-methylethyl)phenyl]amino}-2-oxoethyl)-2-{(2S)-4-methyl-2-[(trifluoroacetyl)amino]pentanoyl}diazanium, CALCIUM ION, Chymotrypsin-like elastase family member 1, ... | | Authors: | Peisach, E, Casebier, D, Gallion, S.L, Furth, P, Petsko, G.A, Hogan Jr, J.C, Ringe, D. | | Deposit date: | 1995-05-01 | | Release date: | 1995-12-07 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Interaction of a peptidomimetic aminimide inhibitor with elastase.
Science, 269, 1995
|
|
3RAT
 
 | |
3B35
 
 | | Crystal structure of the M180A mutant of the aminopeptidase from Vibrio proteolyticus | | Descriptor: | Bacterial leucyl aminopeptidase, SODIUM ION, THIOCYANATE ION, ... | | Authors: | Ataie, N.J, Hoang, Q.Q, Petsko, G.A, Ringe, D. | | Deposit date: | 2007-10-19 | | Release date: | 2007-11-27 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Zinc coordination geometry and ligand binding affinity: the structural and kinetic analysis of the second-shell serine 228 residue and the methionine 180 residue of the aminopeptidase from Vibrio proteolyticus.
Biochemistry, 47, 2008
|
|
3B3T
 
 | | Crystal structure of the D118N mutant of the aminopeptidase from Vibrio proteolyticus | | Descriptor: | Bacterial leucyl aminopeptidase, ISOLEUCINE, SODIUM ION, ... | | Authors: | Ataie, N.J, Hoang, Q.Q, Zahniser, M.P.D, Milne, A, Petsko, G.A, Ringe, D. | | Deposit date: | 2007-10-22 | | Release date: | 2007-11-27 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.17 Å) | | Cite: | Zinc coordination geometry and ligand binding affinity: the structural and kinetic analysis of the second-shell serine 228 residue and the methionine 180 residue of the aminopeptidase from Vibrio proteolyticus.
Biochemistry, 47, 2008
|
|
3AAT
 
 | |
3B3C
 
 | | Crystal structure of the M180A mutant of the aminopeptidase from Vibrio proteolyticus in complex with leucine phosphonic acid | | Descriptor: | Bacterial leucyl aminopeptidase, LEUCINE PHOSPHONIC ACID, POTASSIUM ION, ... | | Authors: | Ataie, N.J, Hoang, Q.Q, Petsko, G.A, Ringe, D. | | Deposit date: | 2007-10-19 | | Release date: | 2007-11-27 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Zinc coordination geometry and ligand binding affinity: the structural and kinetic analysis of the second-shell serine 228 residue and the methionine 180 residue of the aminopeptidase from Vibrio proteolyticus.
Biochemistry, 47, 2008
|
|
3B3S
 
 | | Crystal structure of the M180A mutant of the aminopeptidase from Vibrio proteolyticus in complex with leucine | | Descriptor: | Bacterial leucyl aminopeptidase, LEUCINE, SODIUM ION, ... | | Authors: | Ataie, N.J, Hoang, Q.Q, Petsko, G.A, Ringe, D. | | Deposit date: | 2007-10-22 | | Release date: | 2007-11-27 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.18 Å) | | Cite: | Zinc coordination geometry and ligand binding affinity: the structural and kinetic analysis of the second-shell serine 228 residue and the methionine 180 residue of the aminopeptidase from Vibrio proteolyticus.
Biochemistry, 47, 2008
|
|
