3I6N
 
 | | Mode of Binding of the Tuberculosis Prodrug Isoniazid to Peroxidases: Crystal Structure of Bovine Lactoperoxidase with Isoniazid at 2.7 Resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(DIAZENYLCARBONYL)PYRIDINE, CALCIUM ION, ... | | Authors: | Singh, A.K, Kumar, R.P, Pandey, N, Singh, N, Sinha, M, Bhushan, A, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2009-07-07 | | Release date: | 2009-10-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Mode of binding of the tuberculosis prodrug isoniazid to heme peroxidases: binding studies and crystal structure of bovine lactoperoxidase with isoniazid at 2.7 A resolution.
J.Biol.Chem., 285, 2010
|
|
5ZDK
 
 | | Crystal Structure Analysis of TtQRS in complex with ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CESIUM ION, CHLORIDE ION, ... | | Authors: | Mutharasappan, N, Jain, V, Sharma, A, Manickam, Y, Jeyaraman, J. | | Deposit date: | 2018-02-23 | | Release date: | 2018-12-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structural and functional analysis of Glutaminyl-tRNA synthetase (TtGlnRS) from Thermus thermophilus HB8 and its complexes
Int. J. Biol. Macromol., 120, 2018
|
|
5ZDO
 
 | | Crystal Structure Analysis of TtQRS in co-crystallised with ATP | | Descriptor: | CHLORIDE ION, Glutamine-tRNA ligase | | Authors: | Mutharasappan, N, Jain, V, Sharma, A, Manickam, Y, Jeyaraman, J. | | Deposit date: | 2018-02-23 | | Release date: | 2018-12-05 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural and functional analysis of Glutaminyl-tRNA synthetase (TtGlnRS) from Thermus thermophilus HB8 and its complexes
Int. J. Biol. Macromol., 120, 2018
|
|
5ZDL
 
 | | Crystal Structure Analysis of TtQRS in co-crystallised with ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CHLORIDE ION, Glutamine--tRNA ligase | | Authors: | Mutharasappan, N, Jain, V, Sharma, A, Manickam, Y, Jeyaraman, J. | | Deposit date: | 2018-02-23 | | Release date: | 2018-12-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural and functional analysis of Glutaminyl-tRNA synthetase (TtGlnRS) from Thermus thermophilus HB8 and its complexes
Int. J. Biol. Macromol., 120, 2018
|
|
2JX2
 
 | | Solution conformation of RNA-bound NELF-E RRM | | Descriptor: | Negative elongation factor E | | Authors: | Jampani, N, Schweimer, K, Wenzel, S, Woehrl, B.M, Roesch, P. | | Deposit date: | 2007-11-02 | | Release date: | 2008-04-08 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | NELF-E RRM Undergoes Major Structural Changes in Flexible Protein Regions on Target RNA Binding
Biochemistry, 47, 2008
|
|
7WJL
 
 | | Crystal structure of S. cerevisiae Hos3 | | Descriptor: | ACETATE ION, Histone deacetylase HOS3, ZINC ION | | Authors: | Pang, N.N, Che, S.Y, Yang, N. | | Deposit date: | 2022-01-07 | | Release date: | 2023-01-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural characterization of fungus-specific histone deacetylase Hos3 provides insights into developing selective inhibitors with antifungal activity.
J.Biol.Chem., 298, 2022
|
|
3U1A
 
 | |
3U1C
 
 | |
3U59
 
 | |
7W3D
 
 | | Crystal structure of BRD4 bromodomain 1 (BD1) in complex with N2-(1,2,3-benzotriazol-5-yl)-N3-(dimethylsulfamoyl)-N6-[(2S)-1-methoxypropan-2-yl]pyridine-2,3,6-triamine | | Descriptor: | Bromodomain-containing protein 4, N2-(1,2,3-benzotriazol-5-yl)-N3-(dimethylsulfamoyl)-N6-[(2S)-1-methoxypropan-2-yl]pyridine-2,3,6-triamine | | Authors: | Park, T.H, Lee, B.I. | | Deposit date: | 2021-11-25 | | Release date: | 2022-09-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Discovery of BET specific bromodomain inhibitors with a novel scaffold.
Bioorg.Med.Chem., 72, 2022
|
|
3Q8T
 
 | |
2LIX
 
 | |
7T79
 
 | |
7T78
 
 | |
6F4C
 
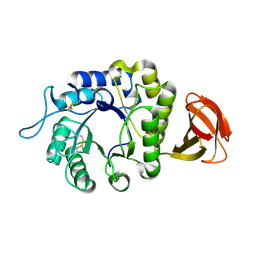 | | Nicotiana benthamiana alpha-galactosidase | | Descriptor: | alpha-galactosidase | | Authors: | Kytidou, K, Aerts, J.M.F.G, Pannu, N.S. | | Deposit date: | 2017-11-29 | | Release date: | 2018-04-25 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Nicotiana benthamianaalpha-galactosidase A1.1 can functionally complement human alpha-galactosidase A deficiency associated with Fabry disease.
J. Biol. Chem., 293, 2018
|
|
2MCG
 
 | |
9F2Q
 
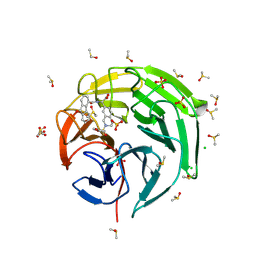 | | Crystal structure of Keap1 kelch domain in complex with a tetrahydroisoquinoline-based small molecule inhibitor at 1.2A resolution | | Descriptor: | (R)-2-(2-((4-methoxyphenyl)sulfonyl)-1,2,3,4-tetrahydroisoquinolin-7-yl)-2-(9-oxo-9H-fluorene-4-carboxamido)acetate, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Narayanan, D, Bach, A, Gajhede, M. | | Deposit date: | 2024-04-23 | | Release date: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structure-Guided Conformational Restriction Leading to High-Affinity, Selective, and Cell-Active Tetrahydroisoquinoline-Based Noncovalent Keap1-Nrf2 Inhibitors.
J.Med.Chem., 2024
|
|
9F2P
 
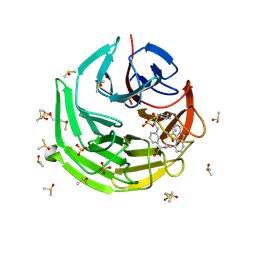 | | Crystal structure of Keap1 kelch domain in complex with a fluorenone-based small molecule inhibitor at 1.36A resolution | | Descriptor: | (R)-2-(3-(((4-methoxyphenyl)sulfonamido)methyl)phenyl)-2-(9-oxo-9H-fluorene-4-carboxamido)acetate, DIMETHYL SULFOXIDE, Kelch-like ECH-associated protein 1, ... | | Authors: | Narayanan, D, Bach, A, Gajhede, M. | | Deposit date: | 2024-04-23 | | Release date: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Structure-Guided Conformational Restriction Leading to High-Affinity, Selective, and Cell-Active Tetrahydroisoquinoline-Based Noncovalent Keap1-Nrf2 Inhibitors.
J.Med.Chem., 2024
|
|
4JIB
 
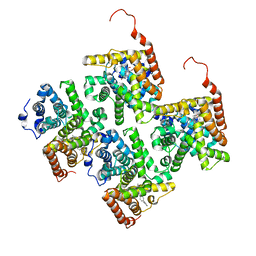 | | Crystal structure of of PDE2-inhibitor complex | | Descriptor: | 1-(2-hydroxyethyl)-3-(2-methylbutan-2-yl)-5-[4-(2-methyl-1H-imidazol-1-yl)phenyl]-6,7-dihydropyrazolo[4,3-e][1,4]diazepin-8(1H)-one, MAGNESIUM ION, ZINC ION, ... | | Authors: | Pandit, J. | | Deposit date: | 2013-03-05 | | Release date: | 2013-05-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Discovery of potent, selective, bioavailable phosphodiesterase 2 (PDE2) inhibitors active in an osteoarthritis pain model, Part I: Transformation of selective pyrazolodiazepinone phosphodiesterase 4 (PDE4) inhibitors into selective PDE2 inhibitors.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
7UHB
 
 | |
7UHC
 
 | |
7OF9
 
 | | Keap1 kelch domain bound to a small molecule fragment | | Descriptor: | DIMETHYL SULFOXIDE, Kelch-like ECH-associated protein 1, [(4S)-6,7-dihydro-4H-thieno[3,2-c]pyran-4-yl]methanamine | | Authors: | Narayanan, D, Bach, A, Gajhede, M. | | Deposit date: | 2021-05-04 | | Release date: | 2022-11-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Development of Noncovalent Small-Molecule Keap1-Nrf2 Inhibitors by Fragment-Based Drug Discovery.
J.Med.Chem., 65, 2022
|
|
7OFD
 
 | | Keap1 kelch domain bound to a small molecule fragment | | Descriptor: | 6-fluoranyl-2-methyl-quinolin-4-ol, DIMETHYL SULFOXIDE, Kelch-like ECH-associated protein 1, ... | | Authors: | Narayanan, D, Bach, A, Gajhede, M. | | Deposit date: | 2021-05-04 | | Release date: | 2022-11-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Development of Noncovalent Small-Molecule Keap1-Nrf2 Inhibitors by Fragment-Based Drug Discovery.
J.Med.Chem., 65, 2022
|
|
7OFA
 
 | | Keap1 kelch domain bound to a small molecule fragment | | Descriptor: | 2,6-bis(chloranyl)-5-fluoranyl-pyridine-3-carboxylic acid, DIMETHYL SULFOXIDE, Kelch-like ECH-associated protein 1 | | Authors: | Narayanan, D, Bach, A, Gajhede, M. | | Deposit date: | 2021-05-04 | | Release date: | 2022-11-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Development of Noncovalent Small-Molecule Keap1-Nrf2 Inhibitors by Fragment-Based Drug Discovery.
J.Med.Chem., 65, 2022
|
|
7OFF
 
 | | Keap1 kelch domain bound to a small molecule inhibitor of the Keap1-Nrf2 protein-protein interaction | | Descriptor: | (2~{R})-2-[(9-oxidanylidenefluoren-4-yl)carbonylamino]-2-phenyl-ethanoic acid, DIMETHYL SULFOXIDE, Kelch-like ECH-associated protein 1, ... | | Authors: | Narayanan, D, Bach, A, Gajhede, M. | | Deposit date: | 2021-05-04 | | Release date: | 2022-11-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | Development of Noncovalent Small-Molecule Keap1-Nrf2 Inhibitors by Fragment-Based Drug Discovery.
J.Med.Chem., 65, 2022
|
|
