5WAM
 
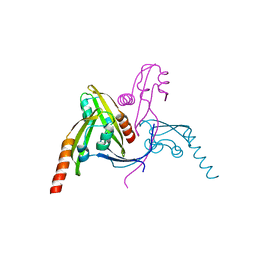 | | Structure of BamE from Neisseria gonorrhoeae | | Descriptor: | Outer membrane protein assembly factor BamE, ZINC ION | | Authors: | Korotkov, K.V, Buchanan, S.K, Noinaj, N. | | Deposit date: | 2017-06-26 | | Release date: | 2017-12-06 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structural and functional insights into the role of BamD and BamE within the beta-barrel assembly machinery in Neisseria gonorrhoeae.
J. Biol. Chem., 293, 2018
|
|
5WAQ
 
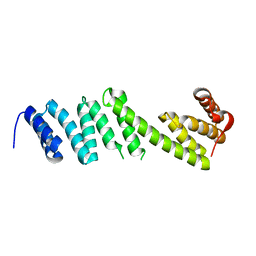 | | Structure of BamD from Neisseria gonorrhoeae | | Descriptor: | Outer membrane protein assembly factor BamD | | Authors: | Korotkov, K.V, Buchanan, S.K, Noinaj, N. | | Deposit date: | 2017-06-26 | | Release date: | 2017-12-06 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.501 Å) | | Cite: | Structural and functional insights into the role of BamD and BamE within the beta-barrel assembly machinery in Neisseria gonorrhoeae.
J. Biol. Chem., 293, 2018
|
|
7K3D
 
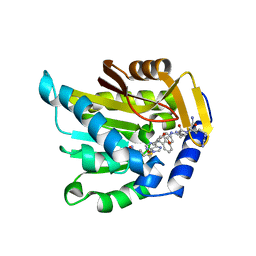 | | The structure of NTMT1 in complex with compound DC1-13 | | Descriptor: | N-terminal Xaa-Pro-Lys N-methyltransferase 1, N~2~-{(2S)-1-[(naphthalen-1-yl)acetyl]-2,5-dihydro-1H-pyrrole-2-carbonyl}-L-lysyl-L-argininamide, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Chen, D, Huang, R, Noinaj, N. | | Deposit date: | 2020-09-11 | | Release date: | 2021-04-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Structure-based Discovery of Cell-Potent Peptidomimetic Inhibitors for Protein N-Terminal Methyltransferase 1.
Acs Med.Chem.Lett., 12, 2021
|
|
7JRK
 
 | |
7JRD
 
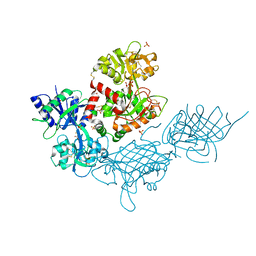 | |
5DYH
 
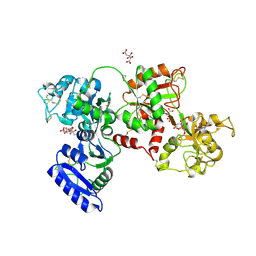 | | Ti(IV) bound human serum transferrin | | Descriptor: | CARBONATE ION, CITRIC ACID, Serotransferrin, ... | | Authors: | Saxena, M, Sharma, S, Noinaj, N, Parks, T.B, Tinoco, A.D. | | Deposit date: | 2015-09-24 | | Release date: | 2016-04-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.682 Å) | | Cite: | Unusual Synergism of Transferrin and Citrate in the Regulation of Ti(IV) Speciation, Transport, and Toxicity.
J.Am.Chem.Soc., 138, 2016
|
|
6OV1
 
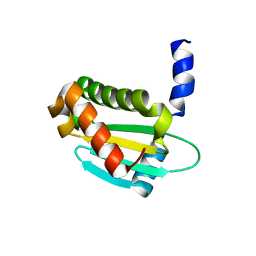 | | Structure of Staphylococcus aureus RNase P protein mutant with defective mRNA degradation activity | | Descriptor: | Ribonuclease P protein component | | Authors: | Ha, L, Colquhoun, J, Noinaj, N, Das, C, Dunman, P, Flaherty, D.P. | | Deposit date: | 2019-05-06 | | Release date: | 2020-12-02 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Genetic and biochemical characterization of Staphylococcus aureus RnpA
To Be Published
|
|
7SOK
 
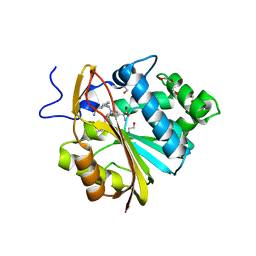 | | Structure of Nicotinamide N-Methyltransferase (NNMT) in complex with inhibitor II329 | | Descriptor: | (2S)-2-amino-4-([3-(3-carbamoylphenyl)prop-2-yn-1-yl]{[(1R,2R,3S,4R)-4-(4-chloro-7H-pyrrolo[2,3-d]pyrimidin-7-yl)-2,3-dihydroxycyclopentyl]methyl}amino)butanoic acid, DI(HYDROXYETHYL)ETHER, NNMT protein | | Authors: | Yadav, R, Iyamu, I.D, Huang, R, Noinaj, N. | | Deposit date: | 2021-10-31 | | Release date: | 2022-11-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Structure of Nicotinamide N-Methyltransferase (NNMT) in complex with inhibitor II329
To Be Published
|
|
7SS1
 
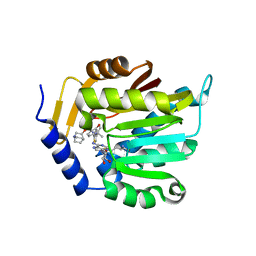 | | The structure of NTMT1 in complex with compound GD433 | | Descriptor: | (1R,3S,4R)-1-azabicyclo[2.2.2]octan-3-yl {2-[2-(4-fluoro-3-hydroxyphenyl)-1,3-thiazol-4-yl]propan-2-yl}carbamate, N-terminal Xaa-Pro-Lys N-methyltransferase 1, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Yadav, R, Guangping, D, Deng, Y, Huang, R, Noinaj, N. | | Deposit date: | 2021-11-09 | | Release date: | 2022-11-16 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Discovery of a first-in-class small molecule inhibitor for Protein N-terminal methyltransferases 1/2
To Be Published
|
|
5SV0
 
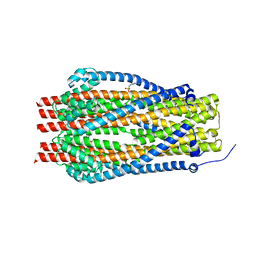 | | Structure of the ExbB/ExbD complex from E. coli at pH 7.0 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Biopolymer transport protein ExbB, CALCIUM ION | | Authors: | Celia, H, Botos, I, Lloubes, R, Buchanan, S.K, Noinaj, N. | | Deposit date: | 2016-08-04 | | Release date: | 2016-09-28 | | Last modified: | 2019-12-11 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural insight into the role of the Ton complex in energy transduction.
Nature, 538, 2016
|
|
5SV1
 
 | | Structure of the ExbB/ExbD complex from E. coli at pH 4.5 | | Descriptor: | Biopolymer transport protein ExbB, Biopolymer transport protein ExbD, MERCURY (II) ION | | Authors: | Celia, H, Botos, I, Lloubes, R, Buchanan, S.K, Noinaj, N. | | Deposit date: | 2016-08-04 | | Release date: | 2016-09-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structural insight into the role of the Ton complex in energy transduction.
Nature, 538, 2016
|
|
8DN7
 
 | |
8DN6
 
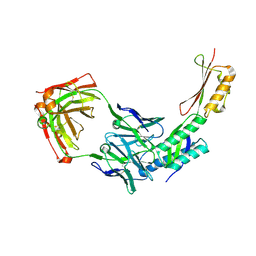 | |
5EKQ
 
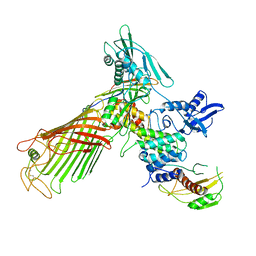 | | The structure of the BamACDE subcomplex from E. coli | | Descriptor: | Outer membrane protein assembly factor BamA, Outer membrane protein assembly factor BamC, Outer membrane protein assembly factor BamD, ... | | Authors: | Bakelar, J, Buchanan, S.K, Noinaj, N. | | Deposit date: | 2015-11-04 | | Release date: | 2016-01-20 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.392 Å) | | Cite: | The structure of the beta-barrel assembly machinery complex.
Science, 351, 2016
|
|
4EXM
 
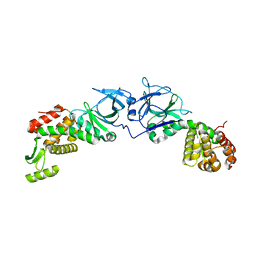 | | The crystal structure of an engineered phage lysin containing the binding domain of pesticin and the killing domain of T4-lysozyme | | Descriptor: | Pesticin, Lysozyme Chimera | | Authors: | Seddiki, N, Noinaj, N, Fairman, J.W, Lukacik, P, Barnard, T.J, Buchanan, S.K. | | Deposit date: | 2012-04-30 | | Release date: | 2012-06-20 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural engineering of a phage lysin that targets Gram-negative pathogens.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4E1S
 
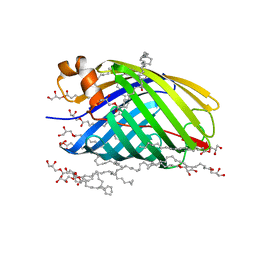 | | X-ray crystal structure of the transmembrane beta-domain from intimin from EHEC strain O157:H7 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2S)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, CHLORIDE ION, ... | | Authors: | Fairman, J.W, Dautin, N, Wojtowicz, D, Wei, L, Noinaj, N, Barnard, T.J, Udho, E, Finkelstein, A, Przytycka, T.M, Cherezov, V, Buchanan, S.K. | | Deposit date: | 2012-03-07 | | Release date: | 2012-06-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.855 Å) | | Cite: | Crystal Structures of the Outer Membrane Domain of Intimin and Invasin from Enterohemorrhagic E. coli and Enteropathogenic Y. pseudotuberculosis.
Structure, 20, 2012
|
|
4E1T
 
 | | X-ray crystal structure of the transmembrane beta-domain from invasin from Yersinia pseudotuberculosis | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2S)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Invasin | | Authors: | Fairman, J.W, Dautin, N, Wojtowicz, D, Wei, L, Noinaj, N, Barnard, T.J, Udho, E, Finkelstein, A, Przytycka, T.M, Cherezov, V, Buchanan, S.K. | | Deposit date: | 2012-03-07 | | Release date: | 2012-06-13 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.263 Å) | | Cite: | Crystal Structures of the Outer Membrane Domain of Intimin and Invasin from Enterohemorrhagic E. coli and Enteropathogenic Y. pseudotuberculosis.
Structure, 20, 2012
|
|
4EPI
 
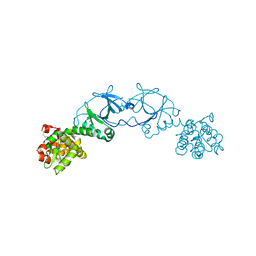 | | The crystal structure of pesticin-T4 lysozyme hybrid stabilized by engineered disulfide bonds | | Descriptor: | Pesticin, Lysozyme Chimera, SODIUM ION, ... | | Authors: | Seddiki, N, Fairman, J.W, Noinaj, N, Lukacik, P, Barnard, T, Buchanan, S.K. | | Deposit date: | 2012-04-17 | | Release date: | 2012-06-20 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Structural engineering of a phage lysin that targets Gram-negative pathogens.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
7RKL
 
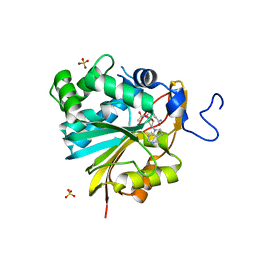 | | Structure of Nicotinamide N-Methyltransferase (NNMT) in complex with II399 (P1 space group) | | Descriptor: | 3-[3-(acetyl{[(1R,2R,3S,4R)-4-(4-chloro-7H-pyrrolo[2,3-d]pyrimidin-7-yl)-2,3-dihydroxycyclopentyl]methyl}amino)prop-1-yn-1-yl]benzamide, NNMT protein, SULFATE ION | | Authors: | Yadav, R, Noinaj, N, Iyamu, I.D, Huang, R. | | Deposit date: | 2021-07-22 | | Release date: | 2022-07-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Exploring Unconventional SAM Analogues To Build Cell-Potent Bisubstrate Inhibitors for Nicotinamide N-Methyltransferase.
Angew.Chem.Int.Ed.Engl., 61, 2022
|
|
7RKK
 
 | | Structure of Nicotinamide N-Methyltransferase (NNMT) in complex with II399 (C2 space group) | | Descriptor: | 3-[3-(acetyl{[(1R,2R,3S,4R)-4-(4-chloro-7H-pyrrolo[2,3-d]pyrimidin-7-yl)-2,3-dihydroxycyclopentyl]methyl}amino)prop-1-yn-1-yl]benzamide, NNMT protein | | Authors: | Yadav, R, Noinaj, N, Iyamu, I.D, Huang, R. | | Deposit date: | 2021-07-22 | | Release date: | 2022-07-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.76 Å) | | Cite: | Exploring Unconventional SAM Analogues To Build Cell-Potent Bisubstrate Inhibitors for Nicotinamide N-Methyltransferase.
Angew.Chem.Int.Ed.Engl., 61, 2022
|
|
3SLJ
 
 | | Pre-cleavage Structure of the Autotransporter EspP - N1023A mutant | | Descriptor: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, Serine protease espP | | Authors: | Barnard, T.B, Noinaj, N, Easley, N.C, Kuszak, A.J, Buchanan, S.K. | | Deposit date: | 2011-06-24 | | Release date: | 2011-11-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.481 Å) | | Cite: | Molecular basis for the activation of a catalytic asparagine residue in a self-cleaving bacterial autotransporter.
J.Mol.Biol., 415, 2012
|
|
8EPI
 
 | | Engineering Crystals with Tunable Symmetries from 14- or 16-Base-Long DNA Strands | | Descriptor: | DNA (5'-D(*CP*GP*AP*CP*G)-3'), DNA (5'-D(P*CP*GP*TP*GP*GP*A)-3'), DNA (5'-D(P*TP*CP*CP*TP*A)-3') | | Authors: | Zhang, C, Zhao, J, Lu, B, Sha, R, Seeman, N.C, Noinaj, N, Mao, C. | | Deposit date: | 2022-10-05 | | Release date: | 2023-03-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Engineering DNA Crystals toward Studying DNA-Guest Molecule Interactions.
J.Am.Chem.Soc., 145, 2023
|
|
8EPE
 
 | | Engineering Crystals with Tunable Symmetries from 14- or 16-Base-Long DNA Strands | | Descriptor: | DNA (5'-D(*CP*GP*AP*CP*G)-3'), DNA (5'-D(P*CP*CP*CP*GP*C)-3'), DNA (5'-D(P*CP*GP*TP*GP*GP*G)-3') | | Authors: | Zhang, C, Zhao, J, Lu, B, Sha, R, Seeman, N.C, Noinaj, N, Mao, C. | | Deposit date: | 2022-10-05 | | Release date: | 2023-03-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Engineering DNA Crystals toward Studying DNA-Guest Molecule Interactions.
J.Am.Chem.Soc., 145, 2023
|
|
8EPB
 
 | | Engineering Crystals with Tunable Symmetries from 14- or 16-Base-Long DNA Strands | | Descriptor: | DNA (5'-D(*CP*GP*CP*TP*GP*GP*TP*GP*GP*TP*TP*CP*GP*A)-3'), DNA (5'-D(*GP*AP*CP*CP*AP*GP*CP*CP*GP*AP*AP*CP*CP*T)-3') | | Authors: | Zhang, C, Zhao, J, Lu, B, Sha, R, Seeman, N.C, Noinaj, N, Mao, C. | | Deposit date: | 2022-10-05 | | Release date: | 2023-03-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Engineering DNA Crystals toward Studying DNA-Guest Molecule Interactions.
J.Am.Chem.Soc., 145, 2023
|
|
8EPD
 
 | | Engineering Crystals with Tunable Symmetries from 14- or 16-Base-Long DNA Strands | | Descriptor: | DNA (5'-D(*CP*GP*AP*CP*G)-3'), DNA (5'-D(P*CP*GP*TP*TP*CP*C)-3'), DNA (5'-D(P*GP*GP*AP*GP*C)-3') | | Authors: | Zhang, C, Zhao, J, Lu, B, Sha, R, Seeman, N.C, Noinaj, N, Mao, C. | | Deposit date: | 2022-10-05 | | Release date: | 2023-03-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Engineering DNA Crystals toward Studying DNA-Guest Molecule Interactions.
J.Am.Chem.Soc., 145, 2023
|
|
