5X3Z
 
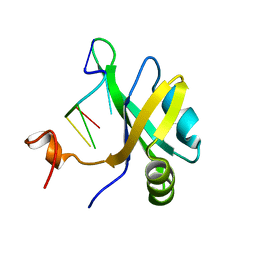 | | Solution structure of musashi1 RBD2 in complex with RNA | | Descriptor: | RNA (5'-R(*GP*UP*AP*GP*U)-3'), RNA-binding protein Musashi homolog 1 | | Authors: | Iwaoka, R, Nagata, T, Tsuda, K, Imai, T, Okano, H, Kobayashi, N, Katahira, M. | | Deposit date: | 2017-02-09 | | Release date: | 2017-12-13 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural Insight into the Recognition of r(UAG) by Musashi-1 RBD2, and Construction of a Model of Musashi-1 RBD1-2 Bound to the Minimum Target RNA
Molecules, 22, 2017
|
|
2RS2
 
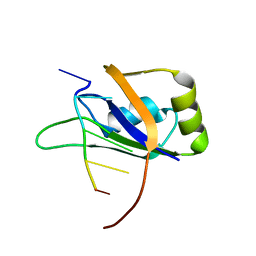 | | 1H, 13C, and 15N Chemical Shift Assignments for Musashi1 RBD1:r(GUAGU) complex | | Descriptor: | RNA (5'-R(*GP*UP*AP*GP*U)-3'), RNA-binding protein Musashi homolog 1 | | Authors: | Ohyama, T, Nagata, T, Tsuda, K, Imai, T, Okano, H, Yamazaki, T, Katahira, M. | | Deposit date: | 2011-06-27 | | Release date: | 2011-12-28 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure of Musashi1 in a complex with target RNA: the role of aromatic stacking interactions
Nucleic Acids Res., 2011
|
|
6K84
 
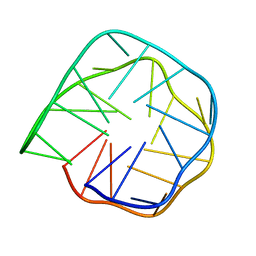 | | Structure of anti-prion RNA aptamer | | Descriptor: | RNA (25-MER) | | Authors: | Mashima, T, Lee, J.H, Hayashi, T, Nagata, T, Kinoshita, M, Katahira, M. | | Deposit date: | 2019-06-11 | | Release date: | 2020-04-01 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Development and structural determination of an anti-PrPCaptamer that blocks pathological conformational conversion of prion protein.
Sci Rep, 10, 2020
|
|
2RU7
 
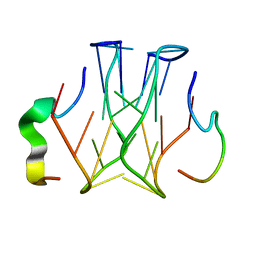 | | Refined structure of RNA aptamer in complex with the partial binding peptide of prion protein | | Descriptor: | P16 peptide from Major prion protein, RNA_(5'-R(*GP*GP*AP*GP*GP*AP*GP*GP*AP*GP*GP*A)-3') | | Authors: | Hayashi, T, Oshima, H, Mashima, T, Nagata, T, Katahira, M, Kinoshita, M. | | Deposit date: | 2013-12-24 | | Release date: | 2014-05-21 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Binding of an RNA aptamer and a partial peptide of a prion protein: crucial importance of water entropy in molecular recognition.
Nucleic Acids Res., 42, 2014
|
|
2RUG
 
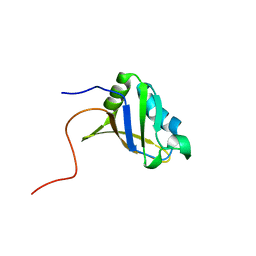 | | Refined solution structure of the first RNA recognition motif domain in CPEB3 | | Descriptor: | Cytoplasmic polyadenylation element-binding protein 3 | | Authors: | Tsuda, K, Kuwasako, K, Nagata, T, Takahashi, M, Kigawa, T, Kobayashi, N, Guntert, P, Shirouzu, M, Yokoyama, S, Muto, Y, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2014-04-15 | | Release date: | 2014-09-17 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Novel RNA recognition motif domain in the cytoplasmic polyadenylation element binding protein 3.
Proteins, 82, 2014
|
|
1WEL
 
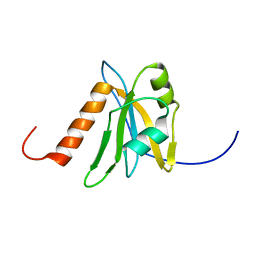 | | Solution structure of RNA binding domain in NP_006038 | | Descriptor: | RNA-binding protein 12 | | Authors: | Someya, T, Muto, Y, Nagata, T, Suzuki, S, Inoue, M, Kigawa, T, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-05-25 | | Release date: | 2005-08-23 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of RNA binding domain in NP_006038
To be Published
|
|
1WG1
 
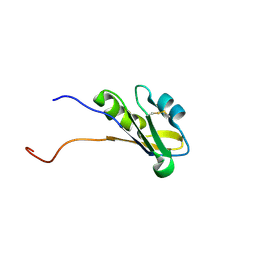 | | Solution structure of RNA binding domain in BAB13405(homolog EXC-7) | | Descriptor: | KIAA1579 protein | | Authors: | Tsuda, K, Muto, Y, Nagata, T, Suzuki, S, Someya, T, Kigawa, T, Terada, T, Shirouzu, M, Inoue, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-05-27 | | Release date: | 2004-11-27 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Solution structure of RNA binding domain in BAB13405(homolog EXC-7)
To be Published
|
|
1WI8
 
 | | Solution structure of the RNA binding domain of eukaryotic initiation factor 4B | | Descriptor: | Eukaryotic translation initiation factor 4B | | Authors: | Suzuki, S, Muto, Y, Nagata, T, Inoue, M, Kigawa, T, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-05-28 | | Release date: | 2004-11-28 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the RNA binding domain of eukaryotic initiation factor 4B
To be Published
|
|
1WI6
 
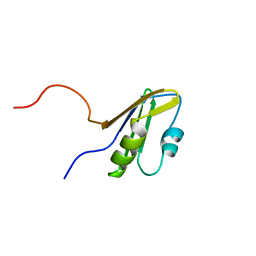 | | Solution structure of the RNA binding domain from mouse hypothetical protein BAB23670 | | Descriptor: | Hypothetical protein (RIKEN cDNA 1300006N24) | | Authors: | Suzuki, S, Muto, Y, Nagata, T, Inoue, M, Kigawa, T, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-05-28 | | Release date: | 2005-06-07 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the RNA binding domain from mouse hypothetical protein BAB23670
To be Published
|
|
6J3G
 
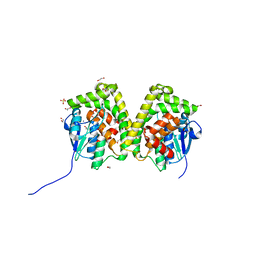 | | Crystal structure of an apo form of the glutathione S-transferase, CsGST83044, of Ceriporiopsis subvermispora | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Glutathione S-transferase, ... | | Authors: | Osman, W.H.W, Mikami, B, Saka, N, Kondo, K, Nagata, T, Katahira, M. | | Deposit date: | 2019-01-04 | | Release date: | 2019-05-15 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Identification of key residues for activities of atypical glutathione S-transferase of Ceriporiopsis subvermispora, a selective degrader of lignin in woody biomass, by crystallography and functional mutagenesis.
Int.J.Biol.Macromol., 132, 2019
|
|
6J3H
 
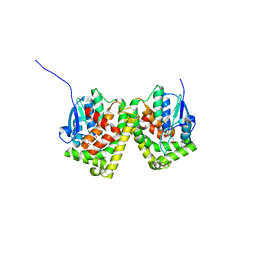 | | Crystal structure of the glutathione S-transferase, CsGST83044, of Ceriporiopsis subvermispora in complex with glutathione | | Descriptor: | GLUTATHIONE, Glutathione S-transferase | | Authors: | Osman, W.H.W, Mikami, B, Saka, N, Kondo, K, Nagata, T, Katahira, M. | | Deposit date: | 2019-01-04 | | Release date: | 2019-05-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Identification of key residues for activities of atypical glutathione S-transferase of Ceriporiopsis subvermispora, a selective degrader of lignin in woody biomass, by crystallography and functional mutagenesis.
Int.J.Biol.Macromol., 132, 2019
|
|
6J3F
 
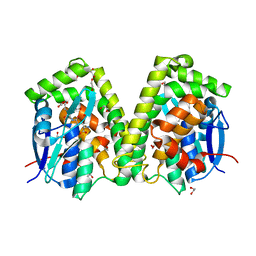 | | Crystal structure of the glutathione S-transferase, CsGST63524, of Ceriporiopsis subvermispora in complex with glutathione | | Descriptor: | 1,2-ETHANEDIOL, GLUTATHIONE, glutathione S-transferase | | Authors: | Osman, W.H.W, Mikami, B, Saka, N, Kondo, K, Nagata, T, Katahira, M. | | Deposit date: | 2019-01-04 | | Release date: | 2019-02-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of a serine-type glutathione S-transferase of Ceriporiopsis subvermispora and identification of the enzymatically important non-canonical residues by functional mutagenesis.
Biochem. Biophys. Res. Commun., 510, 2019
|
|
6J3E
 
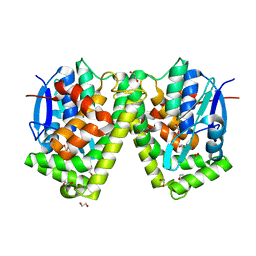 | | Crystal structure of an apo form of the glutathione S-transferase, CsGST63524, of Ceriporiopsis subvermispora | | Descriptor: | 1,2-ETHANEDIOL, glutathione S-transferase | | Authors: | Osman, W.H.W, Mikami, B, Saka, N, Kondo, K, Nagata, T, Katahira, M. | | Deposit date: | 2019-01-04 | | Release date: | 2019-02-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.455 Å) | | Cite: | Structure of a serine-type glutathione S-transferase of Ceriporiopsis subvermispora and identification of the enzymatically important non-canonical residues by functional mutagenesis.
Biochem. Biophys. Res. Commun., 510, 2019
|
|
2RPB
 
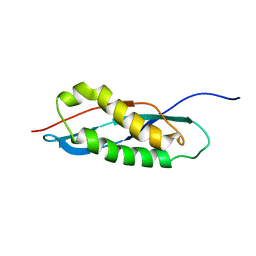 | |
3TK5
 
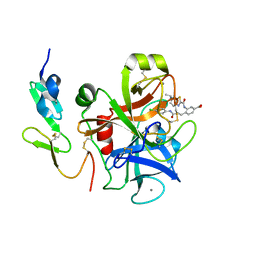 | | Factor Xa in complex with D102-4380 | | Descriptor: | 4-{3-[(4-chlorophenyl)amino]-3-oxopropyl}-3-({[5-(propan-2-yl)-4,5,6,7-tetrahydro[1,3]thiazolo[5,4-c]pyridin-2-yl]carbonyl}amino)benzoic acid, CALCIUM ION, Factor X heavy chain, ... | | Authors: | Suzuki, M, Mochizuki, A, Nagata, T, Takano, H, Kanno, H, Kishida, M, Ohta, T. | | Deposit date: | 2011-08-25 | | Release date: | 2012-08-29 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Zwitter ionic potent durable orally active Factor Xa inhibitor.
To be Published
|
|
3TK6
 
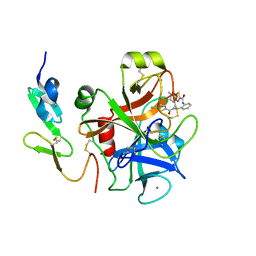 | | factor Xa in complex with D46-5241 | | Descriptor: | CALCIUM ION, Factor X heavy chain, Factor X light chain, ... | | Authors: | Suzuki, M, Mochizuki, A, Nagata, T, Takano, H, Kanno, H, Kishida, M, Ohta, T. | | Deposit date: | 2011-08-25 | | Release date: | 2012-08-29 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Zwitter ionic potent durable orally active Factor Xa inhibitor.
To be Published
|
|
2DNL
 
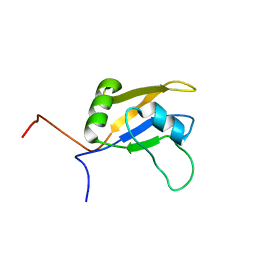 | | Solution structure of RNA binding domain in Cytoplasmic polyadenylation element binding protein 3 | | Descriptor: | cytoplasmic polyadenylation element binding protein 3 | | Authors: | Tsuda, K, Nagata, T, Muto, Y, Inoue, M, Kigawa, T, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-04-26 | | Release date: | 2006-10-26 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of RNA binding domain in Cytoplasmic polyadenylation element binding protein 3
To be Published
|
|
2DNY
 
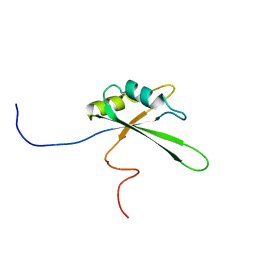 | | Solution structure of the third RNA binding domain of FBP-interacting repressor, SIAHBP1 | | Descriptor: | Fuse-binding protein-interacting repressor, isoform b | | Authors: | Suzuki, S, Nagata, T, Muto, Y, Inoue, M, Kigawa, T, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-04-27 | | Release date: | 2007-04-17 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the third RNA binding domain of FBP-interacting repressor, SIAHBP1
To be Published
|
|
2EI6
 
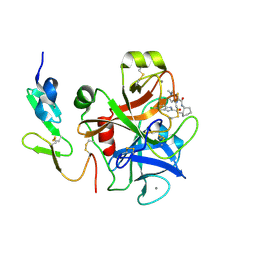 | | FACTOR XA IN COMPLEX WITH THE INHIBITOR (-)-cis-N1-[(5-Chloroindol-2-yl)carbonyl]-N2-[(5-methyl-4,5,6,7-tetrahydrothiazolo[5,4-c]pyridin-2-yl)carbonyl]-1,2-cyclohexanediamine | | Descriptor: | CALCIUM ION, Coagulation factor X, heavy chain, ... | | Authors: | Suzuki, M. | | Deposit date: | 2007-03-12 | | Release date: | 2008-03-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Cycloalkanediamine derivatives as novel blood coagulation factor Xa inhibitors.
Bioorg.Med.Chem.Lett., 17, 2007
|
|
2EI7
 
 | |
2EI8
 
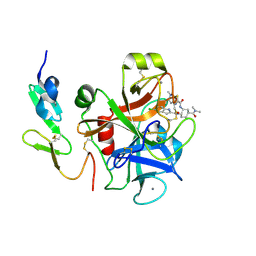 | | FACTOR XA IN COMPLEX WITH THE INHIBITOR (1S,2R,4S)-N1-[(5-chloroindol-2-yl)carbonyl]-4-(N,N-dimethylcarbamoyl)-N2-[(5-methyl-4,5,6,7-tetrahydrothiazolo[5,4-c]pyridin-2-yl)carbonyl]-1,2-cyclohexanediamine | | Descriptor: | CALCIUM ION, Coagulation factor X, heavy chain, ... | | Authors: | Suzuki, M. | | Deposit date: | 2007-03-12 | | Release date: | 2008-03-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Discovery of N-{(1R,2S,5S)-2-{[(5-Chloroindol-2-yl)carbonyl]amino}-5-[(dimethylamino)carbonyl]cyclohexyl}-5-methyl-4,5,6,7-tetrahydrothiazolo[5,4-c]pyridine-2-carboxamide hydrochloride: A Novel, Potent and Orally Active Direct Inhibitor of Factor Xa
To be Published
|
|
3Q3K
 
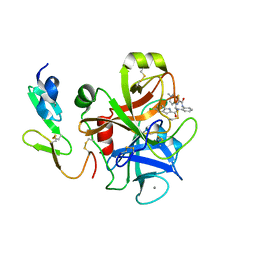 | | Factor Xa in complex with a phenylenediamine derivative | | Descriptor: | Activated factor Xa heavy chain, CALCIUM ION, Factor X light chain, ... | | Authors: | Suzuki, M, Imai, E. | | Deposit date: | 2010-12-22 | | Release date: | 2011-12-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Design, synthesis and SAR of novel ethylenediamine and phenylenediamine derivatives as factor Xa inhibitors.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
1X0F
 
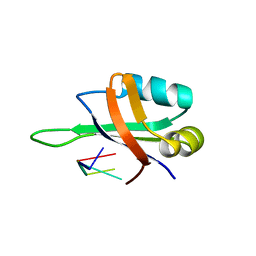 | | Complex structure of the C-terminal RNA-binding domain of hnRNP D(AUF1) with telomeric DNA | | Descriptor: | 5'-D(P*TP*AP*GP*G)-3', Heterogeneous nuclear ribonucleoprotein D0 | | Authors: | Enokizono, Y, Konishi, Y, Nagata, K, Ouhashi, K, Uesugi, S, Ishikawa, F, Katahira, M. | | Deposit date: | 2005-03-22 | | Release date: | 2005-04-05 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structure of hnRNP D complexed with single-stranded telomere DNA and unfolding of the quadruplex by heterogeneous nuclear ribonucleoprotein D.
J.Biol.Chem., 280, 2005
|
|
8H86
 
 | | Cryo-EM structure of the potassium-selective channelrhodopsin HcKCR1 in lipid nanodisc | | Descriptor: | (7R,17E,20E)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSA-17,20-DIEN-1-AMINIUM 4-OXIDE, HcKCR1, PALMITIC ACID, ... | | Authors: | Tajima, S, Kim, Y, Yamashita, K, Fukuda, M, Deisseroth, K, Kato, H.E. | | Deposit date: | 2022-10-21 | | Release date: | 2023-09-06 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (2.56 Å) | | Cite: | Structural basis for ion selectivity in potassium-selective channelrhodopsins.
Cell, 186, 2023
|
|
8H87
 
 | | Cryo-EM structure of the potassium-selective channelrhodopsin HcKCR2 in lipid nanodisc | | Descriptor: | (7R,17E,20E)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSA-17,20-DIEN-1-AMINIUM 4-OXIDE, HcKCR2, PALMITIC ACID, ... | | Authors: | Tajima, S, Kim, Y, Yamashita, K, Fukuda, M, Deisseroth, K, Kato, H.E. | | Deposit date: | 2022-10-21 | | Release date: | 2023-09-06 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (2.53 Å) | | Cite: | Structural basis for ion selectivity in potassium-selective channelrhodopsins.
Cell, 186, 2023
|
|
