7U0E
 
 | |
7U09
 
 | |
7SL5
 
 | |
7K43
 
 | |
7K4N
 
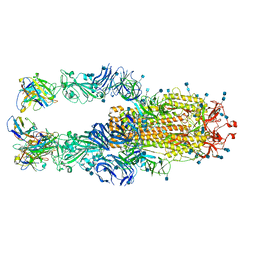 | |
7K45
 
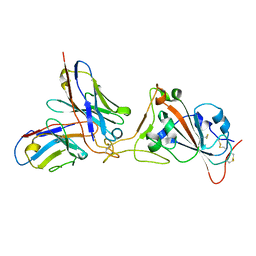 | |
7K3Q
 
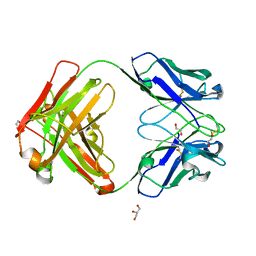 | | An ultra-potent human neutralizing antibody locks the SARS-CoV-2 spike in the closed conformation | | Descriptor: | 1,2-ETHANEDIOL, Fab fragment of S2E12 monoclonal antibody, heavy chain, ... | | Authors: | Snell, G, Czudnochowski, N, Ng, C. | | Deposit date: | 2020-09-12 | | Release date: | 2020-10-07 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Ultrapotent human antibodies protect against SARS-CoV-2 challenge via multiple mechanisms.
Science, 370, 2020
|
|
6V7U
 
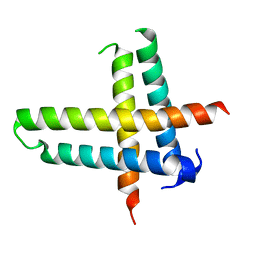 | |
6V7X
 
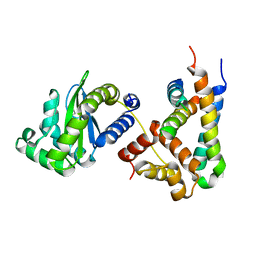 | | Structure of a phage-encoded quorum sensing anti-activator, Aqs1 bound to LasR | | Descriptor: | N-3-OXO-DODECANOYL-L-HOMOSERINE LACTONE, QUORUM SENSING ANTI-ACTIVATOR PROTEIN AQS1, Transcriptional regulator LasR | | Authors: | Shah, M, Moraes, T.F, Maxwell, K.L. | | Deposit date: | 2019-12-09 | | Release date: | 2020-12-16 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | A phage-encoded anti-activator inhibits quorum sensing in Pseudomonas aeruginosa.
Mol.Cell, 81, 2021
|
|
6V7W
 
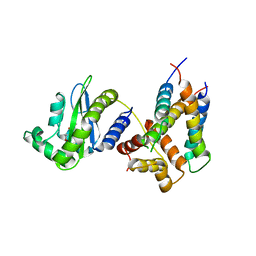 | |
6V7V
 
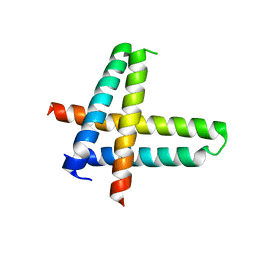 | |
8DYA
 
 | |
8TVB
 
 | |
8TVG
 
 | |
8TVF
 
 | |
8TVE
 
 | |
8TVH
 
 | | Langya henipavirus postfusion F protein in complex with 4G5 Fab, local refinement of the viral membrane proximal region | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4G5 heavy chain, 4G5 light chain, ... | | Authors: | Wang, Z, Seattle Structural Genomics Center for Infectious Disease (SSGCID), Veesler, D. | | Deposit date: | 2023-08-18 | | Release date: | 2024-05-08 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structure and design of Langya virus glycoprotein antigens.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
