1EL5
 
 | | COMPLEX OF MONOMERIC SARCOSINE OXIDASE WITH THE INHIBITOR DIMETHYLGLYCINE | | Descriptor: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, N,N-DIMETHYLGLYCINE, ... | | Authors: | Wagner, M.A, Trickey, P, Chen, Z.-W, Mathews, F.S, Jorns, M.S. | | Deposit date: | 2000-03-13 | | Release date: | 2000-12-06 | | Last modified: | 2017-10-04 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Monomeric sarcosine oxidase: 1. Flavin reactivity and active site binding determinants.
Biochemistry, 39, 2000
|
|
3NBB
 
 | | Crystal structure of mutant Y305F expressed in E. coli in the copper amine oxidase from hansenula polymorpha | | Descriptor: | COPPER (II) ION, Peroxisomal primary amine oxidase | | Authors: | Chen, Z, Datta, S, DuBois, J.L, Klinman, J.P, Mathews, F.S. | | Deposit date: | 2010-06-03 | | Release date: | 2010-08-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Mutation at a strictly conserved, active site tyrosine in the copper amine oxidase leads to uncontrolled oxygenase activity.
Biochemistry, 49, 2010
|
|
1EKM
 
 | | CRYSTAL STRUCTURE AT 2.5 A RESOLUTION OF ZINC-SUBSTITUTED COPPER AMINE OXIDASE OF HANSENULA POLYMORPHA EXPRESSED IN ESCHERICHIA COLI | | Descriptor: | COPPER AMINE OXIDASE, ZINC ION | | Authors: | Chen, Z, Schwartz, B, Williams, N.K, Li, R, Klinman, J.P, Mathews, F.S. | | Deposit date: | 2000-03-09 | | Release date: | 2000-08-23 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure at 2.5 A resolution of zinc-substituted copper amine oxidase of Hansenula polymorpha expressed in Escherichia coli.
Biochemistry, 39, 2000
|
|
1DJN
 
 | | STRUCTURAL AND BIOCHEMICAL CHARACTERIZATION OF RECOMBINANT WILD TYPE TRIMETHYLAMINE DEHYDROGENASE FROM METHYLOPHILUS METHYLOTROPHUS (SP. W3A1) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, FLAVIN MONONUCLEOTIDE, IRON/SULFUR CLUSTER, ... | | Authors: | Trickey, P, Basran, J, Lian, L.-Y, Chen, Z.-W, Barton, J.D, Sutcliffe, M.J, Scrutton, N.S, Mathews, F.S. | | Deposit date: | 1999-12-03 | | Release date: | 1999-12-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and biochemical characterization of recombinant wild type and a C30A mutant of trimethylamine dehydrogenase from methylophilus methylotrophus (sp. W(3)A(1)).
Biochemistry, 39, 2000
|
|
3NBJ
 
 | | Crystal Structure of Y305F mutant of the copper amine oxidase from Hansenula polymorpha expressed in yeast | | Descriptor: | COPPER (II) ION, PHOSPHATE ION, Peroxisomal primary amine oxidase | | Authors: | Chen, Z, Datta, S, DuBois, J.L, Klinman, J.P, Mathews, F.S. | | Deposit date: | 2010-06-03 | | Release date: | 2010-08-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Mutation at a strictly conserved, active site tyrosine in the copper amine oxidase leads to uncontrolled oxygenase activity.
Biochemistry, 49, 2010
|
|
3N9H
 
 | | Crystal Structural of mutant Y305A in the copper amine oxidase from hansenula polymorpha | | Descriptor: | COPPER (II) ION, Peroxisomal primary amine oxidase | | Authors: | Chen, Z, Datta, S, DuBois, J.L, Klinman, J.P, Mathews, F.S. | | Deposit date: | 2010-05-30 | | Release date: | 2010-08-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Mutation at a strictly conserved, active site tyrosine in the copper amine oxidase leads to uncontrolled oxygenase activity.
Biochemistry, 49, 2010
|
|
1G72
 
 | | CATALYTIC MECHANISM OF QUINOPROTEIN METHANOL DEHYDROGENASE: A THEORETICAL AND X-RAY CRYSTALLOGRAPHIC INVESTIGATION | | Descriptor: | CALCIUM ION, METHANOL DEHYDROGENASE HEAVY SUBUNIT, METHANOL DEHYDROGENASE LIGHT SUBUNIT, ... | | Authors: | Zheng, Y, Xia, Z, Chen, Z, Bruice, T.C, Mathews, F.S. | | Deposit date: | 2000-11-08 | | Release date: | 2001-01-24 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Catalytic mechanism of quinoprotein methanol dehydrogenase: A theoretical and x-ray crystallographic investigation.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
1DJQ
 
 | | STRUCTURAL AND BIOCHEMICAL CHARACTERIZATION OF RECOMBINANT C30A MUTANT OF TRIMETHYLAMINE DEHYDROGENASE FROM METHYLOPHILUS METHYLOTROPHUS (SP. W3A1) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, FLAVIN MONONUCLEOTIDE, IRON/SULFUR CLUSTER, ... | | Authors: | Trickey, P, Basran, J, Lian, L.-Y, Chen, Z.-W, Barton, J.D, Sutcliffe, M.J, Scrutton, N.S, Mathews, F.S. | | Deposit date: | 1999-12-03 | | Release date: | 1999-12-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and biochemical characterization of recombinant wild type and a C30A mutant of trimethylamine dehydrogenase from methylophilus methylotrophus (sp. W(3)A(1)).
Biochemistry, 39, 2000
|
|
1DIQ
 
 | | CRYSTAL STRUCTURE OF P-CRESOL METHYLHYDROXYLASE WITH SUBSTRATE BOUND | | Descriptor: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, HEME C, ... | | Authors: | Cunane, L.M, Chen, Z.W, Shamala, N, Mathews, F.S, Cronin, C.S, McIntire, W.S. | | Deposit date: | 1999-11-29 | | Release date: | 1999-12-08 | | Last modified: | 2021-03-03 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structures of the flavocytochrome p-cresol methylhydroxylase and its enzyme-substrate complex: gated substrate entry and proton relays support the proposed catalytic mechanism.
J.Mol.Biol., 295, 2000
|
|
1A2V
 
 | |
1AAN
 
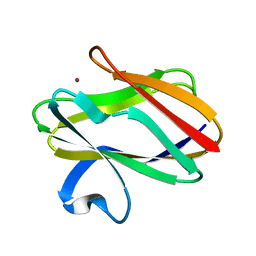 | | CRYSTAL STRUCTURE ANALYSIS OF AMICYANIN AND APOAMICYANIN FROM PARACOCCUS DENITRIFICANS AT 2.0 ANGSTROMS AND 1.8 ANGSTROMS RESOLUTION | | Descriptor: | AMICYANIN, COPPER (II) ION | | Authors: | Chen, L, Durley, R.C.E, Lim, L.W, Mathews, F.S. | | Deposit date: | 1992-04-09 | | Release date: | 1993-10-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure analysis of amicyanin and apoamicyanin from Paracoccus denitrificans at 2.0 A and 1.8 A resolution.
Protein Sci., 2, 1993
|
|
1AAJ
 
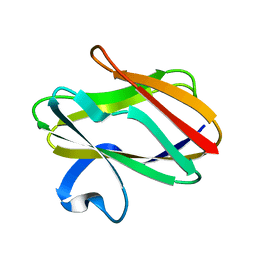 | |
2PGQ
 
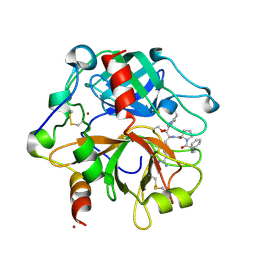 | | Human thrombin mutant C191A-C220A in complex with the inhibitor PPACK | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, D-phenylalanyl-N-[(2S,3S)-6-{[amino(iminio)methyl]amino}-1-chloro-2-hydroxyhexan-3-yl]-L-prolinamide, Thrombin heavy chain, ... | | Authors: | Bush-Pelc, L.A, Marino, F, Chen, Z, Pineda, A.O, Mathews, F.S, Di Cera, E. | | Deposit date: | 2007-04-10 | | Release date: | 2007-07-17 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Important role of the cys-191 cys-220 disulfide bond in thrombin function and allostery
J.Biol.Chem., 282, 2007
|
|
2OCV
 
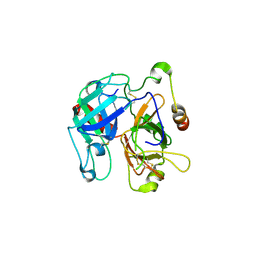 | | Structural basis of Na+ activation mimicry in murine thrombin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Thrombin | | Authors: | Marino, F, Chen, Z, Ergenekan, C.E, Bush, L.A, Mathews, F.S, Di Cera, E. | | Deposit date: | 2006-12-21 | | Release date: | 2007-04-10 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis of na+ activation mimicry in murine thrombin.
J.Biol.Chem., 282, 2007
|
|
2OD3
 
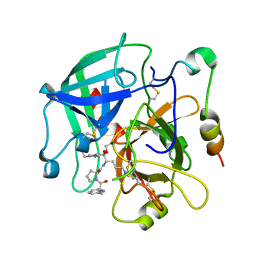 | | Human thrombin chimera with human residues 184a, 186, 186a, 186b, 186c and 222 replaced by murine thrombin equivalents. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, D-phenylalanyl-N-[(2S,3S)-6-{[amino(iminio)methyl]amino}-1-chloro-2-hydroxyhexan-3-yl]-L-prolinamide, Thrombin heavy chain, ... | | Authors: | Marino, F, Chen, Z, Ergenekan, C.E, Bush, L.A, Mathews, F.S, Di Cera, E. | | Deposit date: | 2006-12-21 | | Release date: | 2007-04-10 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural basis of na+ activation mimicry in murine thrombin.
J.Biol.Chem., 282, 2007
|
|
2PGB
 
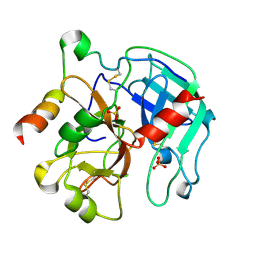 | | Inhibitor-free human thrombin mutant C191A-C220A | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Prothrombin, SULFATE ION | | Authors: | Bush-Pelc, L.A, Marino, F, Chen, Z, Pineda, A.O, Mathews, F.S, Di Cera, E. | | Deposit date: | 2007-04-09 | | Release date: | 2007-07-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Important role of the cys-191 cys-220 disulfide bond in thrombin function and allostery
J.Biol.Chem., 282, 2007
|
|
2GC4
 
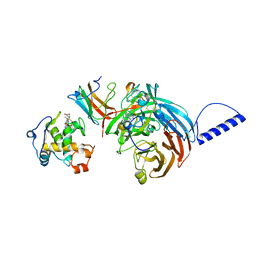 | | Structural comparison of the oxidized ternary electron transfer complex of methylamine dehydrogenase, amicyanin and cytochrome c551i from Paracoccus denitrificans with the substrate-reduced, copper free complex at 1.9 A resolution. | | Descriptor: | Amicyanin, COPPER (II) ION, Cytochrome c-L, ... | | Authors: | Chen, Z, Durley, R, Davidson, V.L, Mathews, F.S. | | Deposit date: | 2006-03-13 | | Release date: | 2006-11-28 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural comparison of the oxidized ternary electron transfer complex of methylamine dehydrogenase, amicyanin and cytochrome c551i from Paracoccus denitrificans with the substrate-reduced, copper free complex at 1.9 A resolution.
To be Published
|
|
2GP9
 
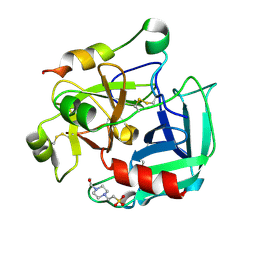 | | Crystal structure of the slow form of thrombin in a self-inhibited conformation | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Prothrombin | | Authors: | Pineda, A, Chen, Z, Mathews, F.S, Di Cera, E. | | Deposit date: | 2006-04-17 | | Release date: | 2006-09-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Crystal structure of thrombin in a self-inhibited conformation.
J.Biol.Chem., 281, 2006
|
|
2GC7
 
 | | Substrate reduced, copper free complex of methylamine dehydrogenase, amicyanin and cytochrome c551i from Paracoccus denitrificans. | | Descriptor: | Amicyanin, Cytochrome c-L, HEME C, ... | | Authors: | Chen, Z, Durley, R, Davidson, V.L, Mathews, F.S. | | Deposit date: | 2006-03-13 | | Release date: | 2007-02-06 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structral comparison of the oxidized ternary electron transfer complex of methylamine dehydrogenase, amicyanin and cytochrome c551i from Paracoccus denitrificans with the substrate-reduced, copper free complex at 1.9 A resolution.
To be Published
|
|
2GF3
 
 | | Structure of the complex of monomeric sarcosine with its substrate analogue inhibitor 2-furoic acid at 1.3 A resolution. | | Descriptor: | 2-FUROIC ACID, CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Chen, Z, Trickey, P, Jorns, M.S, Mathews, F.S. | | Deposit date: | 2006-03-21 | | Release date: | 2007-02-06 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structure of the complex of monomeric sarcosine with its substrate analogue inhibitor 2-furoic acid at 1.3 A resolution.
TO BE PUBLISHED
|
|
1QCW
 
 | | Flavocytochrome B2, ARG289LYS mutant | | Descriptor: | FLAVOCYTOCHROME B2, N-SULFO-FLAVIN MONONUCLEOTIDE | | Authors: | Mowat, C.G, Durley, R.C.E, Pike, A.D, Barton, J.D, Chen, Z.-W, Mathews, F.S, Lederer, F, Reid, G.A, Chapman, S.K. | | Deposit date: | 1999-05-07 | | Release date: | 1999-05-24 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Kinetic and crystallographic studies on the active site Arg289Lys mutant of flavocytochrome b2 (yeast L-lactate dehydrogenase)
Biochemistry, 39, 2000
|
|
1KBI
 
 | | Crystallographic Study of the Recombinant Flavin-binding Domain of Baker's Yeast Flavocytochrome b2: Comparison with the Intact Wild-type Enzyme | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CYTOCHROME B2, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Cunane, L.M, Barton, J.D, Chen, Z.-W, Welsh, F.E, Chapman, S.K, Reid, G.A, Mathews, F.S. | | Deposit date: | 2001-11-06 | | Release date: | 2002-04-03 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystallographic study of the recombinant flavin-binding domain of Baker's yeast flavocytochrome b(2): comparison with the intact wild-type enzyme.
Biochemistry, 41, 2002
|
|
1PBY
 
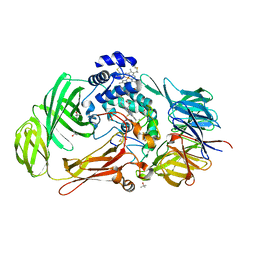 | | Structure of the Phenylhydrazine Adduct of the Quinohemoprotein Amine Dehydrogenase from Paracoccus denitrificans at 1.7 A Resolution | | Descriptor: | HEME C, TERTIARY-BUTYL ALCOHOL, quinohemoprotein amine dehydrogenase 40 kDa subunit, ... | | Authors: | Datta, S, Ikeda, T, Kano, K, Mathews, F.S. | | Deposit date: | 2003-05-15 | | Release date: | 2003-09-02 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of the phenylhydrazine adduct of the quinohemoprotein amine dehydrogenase from Paracoccus denitrificans at 1.7 A resolution.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
1KBJ
 
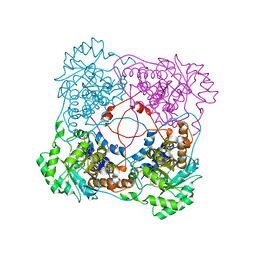 | | Crystallographic Study of the Recombinant Flavin-binding Domain of Baker's Yeast Flavocytochrome b2: comparison with the Intact Wild-type Enzyme | | Descriptor: | 1,2-ETHANEDIOL, CYTOCHROME B2, FLAVIN MONONUCLEOTIDE | | Authors: | Cunane, L.M, Barton, J.D, Chen, Z.W, Welsh, F.E, Chapman, S.K, Reid, G.A, Mathews, F.S. | | Deposit date: | 2001-11-06 | | Release date: | 2002-04-03 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystallographic study of the recombinant flavin-binding domain of Baker's yeast flavocytochrome b(2): comparison with the intact wild-type enzyme.
Biochemistry, 41, 2002
|
|
1KV9
 
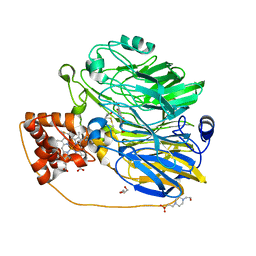 | | Structure at 1.9 A Resolution of a Quinohemoprotein Alcohol Dehydrogenase from Pseudomonas putida HK5 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ACETONE, CALCIUM ION, ... | | Authors: | Chen, Z.-W, Matsushita, K, Yamashita, T, Fujii, T, Toyama, H, Adachi, O, Bellamy, H.D, Mathews, F.S. | | Deposit date: | 2002-01-25 | | Release date: | 2002-07-10 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure at 1.9 A resolution of a quinohemoprotein alcohol dehydrogenase from Pseudomonas putida HK5.
Structure, 10, 2002
|
|
