6ASQ
 
 | | Structure of Grp94 bound to methyl 2-[2-(2-benzylpyridin-3-yl)ethyl]-3-chloro-4,6-dihydroxybenzoate, a pan-Hsp90 inhibitor | | Descriptor: | 3,6,9,12,15,18,21-HEPTAOXATRICOSANE-1,23-DIOL, DI(HYDROXYETHYL)ETHER, Endoplasmin, ... | | Authors: | Huard, D.J.E, Lieberman, R.L. | | Deposit date: | 2017-08-25 | | Release date: | 2018-04-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Trifunctional High-Throughput Screen Identifies Promising Scaffold To Inhibit Grp94 and Treat Myocilin-Associated Glaucoma.
ACS Chem. Biol., 13, 2018
|
|
4WXQ
 
 | | Crystal Structure of the Myocilin Olfactomedin Domain | | Descriptor: | CALCIUM ION, HEXAETHYLENE GLYCOL, Myocilin, ... | | Authors: | Orwig, S.D, Turnage, K.C, Donegan, R.K, Lieberman, R.L. | | Deposit date: | 2014-11-14 | | Release date: | 2015-04-01 | | Last modified: | 2017-09-06 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural basis for misfolding in myocilin-associated glaucoma.
Hum.Mol.Genet., 24, 2015
|
|
4XAV
 
 | | Crystal structure of olfactomedin domain from gliomedin | | Descriptor: | GLYCEROL, Gliomedin, PHOSPHATE ION, ... | | Authors: | Hill, S.E, Nguyen, E, Lieberman, R.L. | | Deposit date: | 2014-12-15 | | Release date: | 2015-07-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.052 Å) | | Cite: | Molecular Details of Olfactomedin Domains Provide Pathway to Structure-Function Studies.
Plos One, 10, 2015
|
|
4XAT
 
 | | Crystal structure of the olfactomedin domain from noelin/pancortin/olfactomedin-1 | | Descriptor: | CALCIUM ION, GLYCEROL, Noelin, ... | | Authors: | Hill, S.E, Nguyen, E, Lieberman, R.L. | | Deposit date: | 2014-12-15 | | Release date: | 2015-07-15 | | Last modified: | 2022-03-30 | | Method: | X-RAY DIFFRACTION (2.113 Å) | | Cite: | Molecular Details of Olfactomedin Domains Provide Pathway to Structure-Function Studies.
Plos One, 10, 2015
|
|
4WXS
 
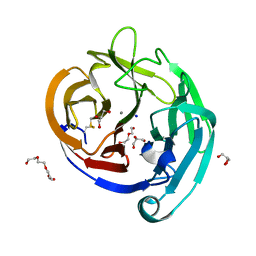 | | Crystal Structure of the E396D SNP Variant of the Myocilin Olfactomedin Domain | | Descriptor: | CALCIUM ION, GLYCEROL, HEXAETHYLENE GLYCOL, ... | | Authors: | Donegan, R.K, Freeman, D.M, Lieberman, R.L. | | Deposit date: | 2014-11-14 | | Release date: | 2015-04-01 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for misfolding in myocilin-associated glaucoma.
Hum.Mol.Genet., 24, 2015
|
|
4X0K
 
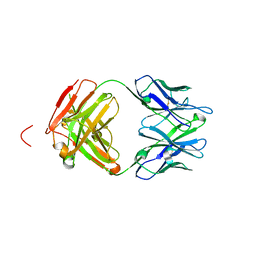 | | Engineered Fab fragment specific for EYMPME (EE) peptide | | Descriptor: | Fab fragment heavy chain, Fab fragment light chain | | Authors: | Johnson, J.L, Lieberman, R.L. | | Deposit date: | 2014-11-21 | | Release date: | 2015-04-08 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Structural and biophysical characterization of an epitope-specific engineered Fab fragment and complexation with membrane proteins: implications for co-crystallization.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4NKM
 
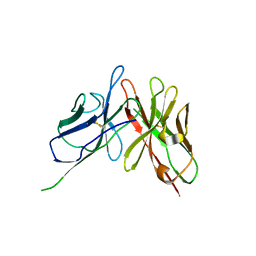 | | Crystal structure of engineered anti-EE scFv antibody fragment | | Descriptor: | Engineered scFv | | Authors: | Kalyoncu, S, Hyun, J, Pai, J.C, Johnson, J.L, Etzminger, K, Jain, A, Heaner Jr, D, Molares, I.A, Truskett, T.M, Maynard, J.A, Lieberman, R.L. | | Deposit date: | 2013-11-12 | | Release date: | 2014-03-12 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3.71 Å) | | Cite: | Effects of protein engineering and rational mutagenesis on crystal lattice of single chain antibody fragments.
Proteins, 82, 2014
|
|
4NKD
 
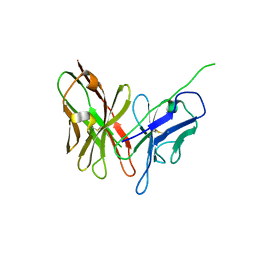 | | Crystal structure of engineered anti-EE scFv antibody fragment | | Descriptor: | Engineered scFv | | Authors: | Kalyoncu, S, Hyun, J, Pai, J.C, Johnson, J.L, Etzminger, K, Jain, A, Heaner Jr, D, Molares, I.A, Truskett, T.M, Maynard, J.A, Lieberman, R.L. | | Deposit date: | 2013-11-12 | | Release date: | 2014-03-12 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3.303 Å) | | Cite: | Effects of protein engineering and rational mutagenesis on crystal lattice of single chain antibody fragments.
Proteins, 82, 2014
|
|
4NKO
 
 | | Crystal structure of engineered anti-EE scFv antibody fragment | | Descriptor: | Engineered scFv | | Authors: | Kalyoncu, S, Hyun, J, Pai, J.C, Johnson, J.L, Etzminger, K, Jain, A, Heaner Jr, D, Molares, I.A, Truskett, T.M, Maynard, J.A, Lieberman, R.L. | | Deposit date: | 2013-11-12 | | Release date: | 2014-03-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.496 Å) | | Cite: | Effects of protein engineering and rational mutagenesis on crystal lattice of single chain antibody fragments.
Proteins, 82, 2014
|
|
4NXS
 
 | | Crystal structure of human alpha-galactosidase A in complex with 1-deoxygalactonojirimycin-pFPhT | | Descriptor: | (2R,3S,4R,5S)-N-(4-fluorophenyl)-3,4,5-trihydroxy-2-(hydroxymethyl)piperidine-1-carbothioamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Johnson, J.L, Drury, J.E, Lieberman, R.L. | | Deposit date: | 2013-12-09 | | Release date: | 2014-06-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.5493 Å) | | Cite: | Molecular Basis of 1-Deoxygalactonojirimycin Arylthiourea Binding to Human alpha-Galactosidase A: Pharmacological Chaperoning Efficacy on Fabry Disease Mutants.
Acs Chem.Biol., 9, 2014
|
|
5U7E
 
 | |
5U7F
 
 | |
5U7H
 
 | |
6OU3
 
 | |
6OU0
 
 | |
6OU1
 
 | | Crystal Structure of the Computationally-derived 21-Variant of the Myocilin Olfactomedin Domain | | Descriptor: | CALCIUM ION, GLYCEROL, Myocilin, ... | | Authors: | Hill, S.E, Kwon, M.S, Lieberman, R.L. | | Deposit date: | 2019-05-03 | | Release date: | 2019-07-03 | | Last modified: | 2020-01-15 | | Method: | X-RAY DIFFRACTION (1.878 Å) | | Cite: | Stable calcium-free myocilin olfactomedin domain variants reveal challenges in differentiating between benign and glaucoma-causing mutations.
J.Biol.Chem., 294, 2019
|
|
6OU2
 
 | |
5K8N
 
 | | 5NAA-bound 5-nitroanthranilate aminohydrolase | | Descriptor: | 5-nitroanthranilic acid, 5-nitroanthranilic acid aminohydrolase | | Authors: | Kalyoncu, S. | | Deposit date: | 2016-05-30 | | Release date: | 2016-10-05 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.225 Å) | | Cite: | Enzymatic hydrolysis by transition-metal-dependent nucleophilic aromatic substitution.
Nat.Chem.Biol., 12, 2016
|
|
5K8M
 
 | | Apo 5-nitroanthranilate aminohydrolase | | Descriptor: | 5-nitroanthranilic acid aminohydrolase | | Authors: | Kalyoncu, S. | | Deposit date: | 2016-05-30 | | Release date: | 2016-10-05 | | Last modified: | 2016-11-23 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Enzymatic hydrolysis by transition-metal-dependent nucleophilic aromatic substitution.
Nat.Chem.Biol., 12, 2016
|
|
5K8P
 
 | | Zn2+/Tetrahedral intermediate-bound R289A 5-nitroanthranilate aminohydrolase | | Descriptor: | (6~{R})-6-azanyl-3-nitro-6-oxidanyl-cyclohexa-1,3-diene-1-carboxylic acid, 5-nitroanthranilic acid aminohydrolase, GLYCEROL, ... | | Authors: | Kalyoncu, S. | | Deposit date: | 2016-05-30 | | Release date: | 2016-10-05 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Enzymatic hydrolysis by transition-metal-dependent nucleophilic aromatic substitution.
Nat.Chem.Biol., 12, 2016
|
|
5K8O
 
 | | Mn2+/5NSA-bound 5-nitroanthranilate aminohydrolase | | Descriptor: | 5-nitroanthranilic acid aminohydrolase, 5-nitrosalicylic acid, MANGANESE (II) ION | | Authors: | Kalyoncu, S. | | Deposit date: | 2016-05-30 | | Release date: | 2016-10-05 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.893 Å) | | Cite: | Enzymatic hydrolysis by transition-metal-dependent nucleophilic aromatic substitution.
Nat.Chem.Biol., 12, 2016
|
|
