7X9D
 
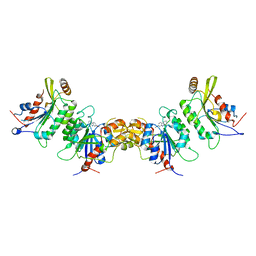 | | DNMT3B in complex with harmine | | Descriptor: | 7-METHOXY-1-METHYL-9H-BETA-CARBOLINE, DNA (cytosine-5)-methyltransferase 3-like, DNA (cytosine-5)-methyltransferase 3B | | Authors: | Cho, C.-C, Yuan, H.S. | | Deposit date: | 2022-03-15 | | Release date: | 2023-03-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.08 Å) | | Cite: | Mechanistic Insights into Harmine-Mediated Inhibition of Human DNA Methyltransferases and Prostate Cancer Cell Growth.
Acs Chem.Biol., 18, 2023
|
|
6XRJ
 
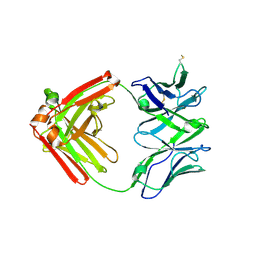 | |
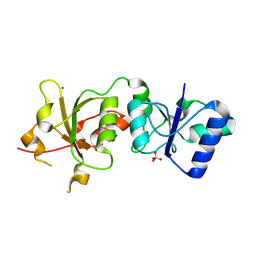
3PXA
 
 | |
3PXD
 
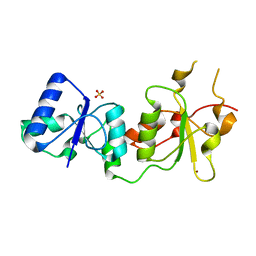 | |
3PXE
 
 | |
3PXB
 
 | |
3PXC
 
 | |
2ZX0
 
 | | Rhamnose-binding lectin CSL3 | | Descriptor: | CSL3, GLYCEROL, PHOSPHATE ION | | Authors: | Shirai, T, Watababe, Y, Lee, M, Ogawa, T, Muramoto, K. | | Deposit date: | 2008-12-19 | | Release date: | 2009-06-30 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of rhamnose-binding lectin CSL3: unique pseudo-tetrameric architecture of a pattern recognition protein
J.Mol.Biol., 391, 2009
|
|
2ZX2
 
 | | Rhamnose-binding lectin CSL3 | | Descriptor: | CSL3, PHOSPHATE ION, alpha-L-rhamnopyranose | | Authors: | Shirai, T, Watababe, Y, Lee, M, Ogawa, T, Muramoto, K. | | Deposit date: | 2008-12-19 | | Release date: | 2009-06-30 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of rhamnose-binding lectin CSL3: unique pseudo-tetrameric architecture of a pattern recognition protein
J.Mol.Biol., 391, 2009
|
|
2ZX1
 
 | | Rhamnose-binding lectin CSL3 | | Descriptor: | CSL3, PHOSPHATE ION | | Authors: | Shirai, T, Watababe, Y, Lee, M, Ogawa, T, Muramoto, K. | | Deposit date: | 2008-12-19 | | Release date: | 2009-06-30 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of rhamnose-binding lectin CSL3: unique pseudo-tetrameric architecture of a pattern recognition protein
J.Mol.Biol., 391, 2009
|
|
2ORY
 
 | | Crystal structure of M37 lipase | | Descriptor: | Lipase | | Authors: | Jung, S.K, Jeong, D.G, Kim, S.J. | | Deposit date: | 2007-02-05 | | Release date: | 2008-01-29 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis for the cold adaptation of psychrophilic M37 lipase from Photobacterium lipolyticum.
Proteins, 71, 2008
|
|
6A07
 
 | |
6A02
 
 | |
6A09
 
 | | Salmonella Typhi YfdX in the P222 space group | | Descriptor: | YfdX protein | | Authors: | Ku, B, Lee, H.S, Kim, S.J. | | Deposit date: | 2018-06-05 | | Release date: | 2019-02-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.293 Å) | | Cite: | Structural and Physiological Exploration ofSalmonellaTyphi YfdX Uncovers Its Dual Function in Bacterial Antibiotic Stress and Virulence.
Front Microbiol, 9, 2018
|
|
2L65
 
 | | HADDOCK calculated model of the complex of the resistance protein CalC and Calicheamicin-Gamma | | Descriptor: | 2,4-dideoxy-4-(ethylamino)-3-O-methyl-alpha-L-threo-pentopyranose-(1-2)-4-amino-4,6-dideoxy-beta-D-glucopyranose, 2,6-dideoxy-4-thio-beta-D-allopyranose, 3-O-methyl-alpha-L-rhamnopyranose, ... | | Authors: | Singh, S, Markley, J.L, Thorson, J.S, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2010-11-15 | | Release date: | 2011-03-02 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural insight into the self-sacrifice mechanism of enediyne resistance.
Acs Chem.Biol., 1, 2006
|
|
3NPS
 
 | |
2EVN
 
 | | NMR solution structures of At1g77540 | | Descriptor: | Protein At1g77540 | | Authors: | Tyler, R.C, Singh, S, Tonelli, M, Min, M.S, Markley, J.L, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2005-10-31 | | Release date: | 2005-11-15 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of Arabidopsis thaliana At1g77540 Protein, a Minimal Acetyltransferase from the COG2388 Family.
Biochemistry, 45, 2006
|
|
4ANV
 
 | | Complexes of PI3Kgamma with isoform selective inhibitors. | | Descriptor: | 2-{4-[(4'-METHOXYBIPHENYL-3-YL)SULFONYL]PIPERAZIN-1-YL}-3-(4-METHOXYPHENYL)PYRAZINE, PHOSPHATIDYLINOSITOL-4,5-BISPHOSPHATE 3-KINASE CATALYTIC SUBUNIT GAMMA ISOFORM, SULFATE ION | | Authors: | Foster, P.G, Lougheed, J.C. | | Deposit date: | 2012-03-22 | | Release date: | 2012-05-09 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | The Discovery of a Novel Series of Potent and Orally Bioavailable Phosphoinositide 3-Kinase Gamma Inhibitors
J.Med.Chem., 55, 2012
|
|
4ANX
 
 | | Complexes of PI3Kgamma with isoform selective inhibitors. | | Descriptor: | 5-{3-[(4-{3-[4-(1-methylethyl)phenyl]pyrazin-2-yl}piperazin-1-yl)sulfonyl]phenyl}pyrimidin-2-amine, PHOSPHATIDYLINOSITOL-4,5-BISPHOSPHATE 3-KINASE CATALYTIC SUBUNIT GAMMA ISOFORM | | Authors: | Foster, P.G, Lougheed, J.C. | | Deposit date: | 2012-03-22 | | Release date: | 2012-05-09 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | The Discovery of a Novel Series of Potent and Orally Bioavailable Phosphoinositide 3-Kinase Gamma Inhibitors
J.Med.Chem., 55, 2012
|
|
4BBE
 
 | | Aminoalkylpyrimidine Inhibitor Complexes with JAK2 | | Descriptor: | N-[4-[2-[(4-morpholin-4-ylphenyl)amino]pyrimidin-4-yl]phenyl]ethanamide, TYROSINE-PROTEIN KINASE JAK2 | | Authors: | Li, J. | | Deposit date: | 2012-09-21 | | Release date: | 2012-11-21 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Sar and in Vivo Evaluation of 4-Aryl-2-Aminoalkylpyrimidines as Potent and Selective Janus Kinase 2 (Jak2) Inhibitors
Bioorg.Med.Chem.Lett., 22, 2012
|
|
4BBF
 
 | | Aminoalkylpyrimidine Inhibitor Complexes with JAK2 | | Descriptor: | (2R)-N-[4-[2-[(4-morpholin-4-ylphenyl)amino]pyrimidin-4-yl]phenyl]pyrrolidine-2-carboxamide, TYROSINE-PROTEIN KINASE JAK2 | | Authors: | Li, J. | | Deposit date: | 2012-09-21 | | Release date: | 2012-11-21 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Sar and in Vivo Evaluation of 4-Aryl-2-Aminoalkylpyrimidines as Potent and Selective Janus Kinase 2 (Jak2) Inhibitors
Bioorg.Med.Chem.Lett., 22, 2012
|
|
4ANU
 
 | | Complexes of PI3Kgamma with isoform selective inhibitors. | | Descriptor: | 3-AMINO-N-METHYL-6-[3-(1H-TETRAZOL-5-YL)PHENYL]PYRAZINE-2-CARBOXAMIDE, PHOSPHATIDYLINOSITOL-4,5-BISPHOSPHATE 3-KINASE CATALYTIC SUBUNIT GAMMA ISOFORM, SULFATE ION | | Authors: | Foster, P.G, Lougheed, J.C. | | Deposit date: | 2012-03-22 | | Release date: | 2012-05-09 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | The Discovery of a Novel Series of Potent and Orally Bioavailable Phosphoinositide 3-Kinase Gamma Inhibitors
J.Med.Chem., 55, 2012
|
|
4ANW
 
 | | Complexes of PI3Kgamma with isoform selective inhibitors. | | Descriptor: | 3-AMINO-6-{4-CHLORO-3-[(2,3-DIFLUOROPHENYL)SULFAMOYL]PHENYL}-N-METHYLPYRAZINE-2-CARBOXAMIDE, PHOSPHATIDYLINOSITOL-4,5-BISPHOSPHATE 3-KINASE CATALYTIC SUBUNIT GAMMA ISOFORM, SULFATE ION | | Authors: | Foster, P.G, Lougheed, J.C. | | Deposit date: | 2012-03-22 | | Release date: | 2012-05-09 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | The Discovery of a Novel Series of Potent and Orally Bioavailable Phosphoinositide 3-Kinase Gamma Inhibitors
J.Med.Chem., 55, 2012
|
|
6CBP
 
 | |
1VJH
 
 | | Crystal structure of gene product of At1g24000 from Arabidopsis thaliana | | Descriptor: | Bet v I allergen family | | Authors: | Wesenberg, G.E, Smith, D.W, Phillips Jr, G.N, Johnson, K.A, Bingman, C.A, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2004-02-20 | | Release date: | 2004-03-16 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | 1H, 15N and 13C resonance assignments of the putative Bet v 1 family protein At1g24000.1 from Arabidopsis thaliana.
J.Biomol.Nmr, 32, 2005
|
|
