7FER
 
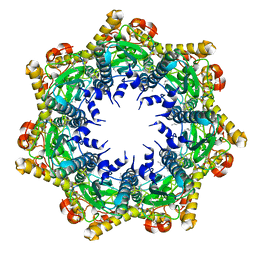 | | Cryo-EM structure of BsClpP-ADEP1 complex at pH 4.2 | | Descriptor: | ADEP1, ATP-dependent Clp protease proteolytic subunit | | Authors: | Kim, L, Lee, B.-G, Kim, M.K, Kwon, D.H, Kim, H, Brotz-Oesterhelt, H, Roh, S.-H, Song, H.K. | | Deposit date: | 2021-07-21 | | Release date: | 2022-07-06 | | Last modified: | 2022-07-20 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural insights into ClpP protease side exit pore-opening by a pH drop coupled with substrate hydrolysis.
Embo J., 41, 2022
|
|
8I9Q
 
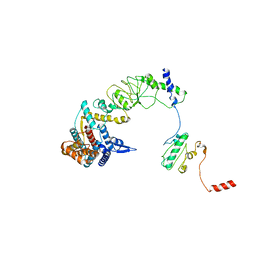 | | The focused refinement of CCT4-PhLP2A from TRiC-PhLP2A complex in the open state | | Descriptor: | Phosducin-like protein 3, T-complex protein 1 subunit delta | | Authors: | Roh, S.H, Park, J, Kim, H, Lim, S. | | Deposit date: | 2023-02-07 | | Release date: | 2023-12-20 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (4.22 Å) | | Cite: | A structural vista of phosducin-like PhLP2A-chaperonin TRiC cooperation during the ATP-driven folding cycle.
Nat Commun, 15, 2024
|
|
8IB8
 
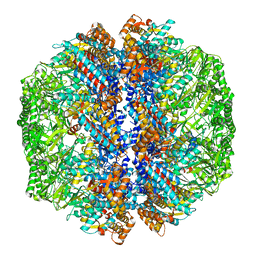 | | Human TRiC-PhLP2A-actin complex in the closed state | | Descriptor: | ACTB protein (Fragment), Phosducin-like protein 3, T-complex protein 1 subunit alpha, ... | | Authors: | Roh, S.H, Park, J, Kim, H, Lim, S. | | Deposit date: | 2023-02-09 | | Release date: | 2023-12-20 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (4.42 Å) | | Cite: | A structural vista of phosducin-like PhLP2A-chaperonin TRiC cooperation during the ATP-driven folding cycle.
Nat Commun, 15, 2024
|
|
8I9U
 
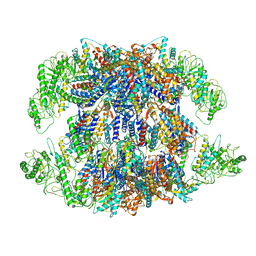 | | Human TRiC-PhLP2A complex in the open state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Phosducin-like protein 3, T-complex protein 1 subunit alpha, ... | | Authors: | Roh, S.H, Park, J, Kim, H, Lim, S. | | Deposit date: | 2023-02-07 | | Release date: | 2024-01-17 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | A structural vista of phosducin-like PhLP2A-chaperonin TRiC cooperation during the ATP-driven folding cycle.
Nat Commun, 15, 2024
|
|
8I1U
 
 | | Human TRiC-PhLP2A complex in the closed state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, MAGNESIUM ION, ... | | Authors: | Roh, S.H, Park, J, Kim, H, Lim, S. | | Deposit date: | 2023-01-13 | | Release date: | 2024-01-31 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.24 Å) | | Cite: | A structural vista of phosducin-like PhLP2A-chaperonin TRiC cooperation during the ATP-driven folding cycle.
Nat Commun, 15, 2024
|
|
8I6J
 
 | | The focused refinement of CCT3-PhLP2A from TRiC-PhLP2A complex in the open state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Phosducin-like protein 3, T-complex protein 1 subunit gamma | | Authors: | Roh, S.H, Park, J, Kim, H, Lim, S. | | Deposit date: | 2023-01-28 | | Release date: | 2024-01-31 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.82 Å) | | Cite: | A structural vista of phosducin-like PhLP2A-chaperonin TRiC cooperation during the ATP-driven folding cycle.
Nat Commun, 15, 2024
|
|
9J03
 
 | | Cyro-EM Structure of Human TLR4/MD-2/DLAM1 Complex | | Descriptor: | (3R)-3-(dodecanoyloxy)tetradecanoic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Fu, Y, Kim, H, Zamyatina, A, Kim, H.M. | | Deposit date: | 2024-08-02 | | Release date: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structural insight into TLR4/MD-2 activation by synthetic LPS mimetics with distinct binding modes.
Nat Commun, 16, 2025
|
|
8KHO
 
 | | Crystal structure of human methionine aminopeptidase 12 (MAP12) in complex with two Cobalt ions and Methionine | | Descriptor: | COBALT (II) ION, METHIONINE, Methionine aminopeptidase 1D, ... | | Authors: | Lee, Y, Lee, E, Hahn, H, Kim, H, Heo, Y, Jang, D.M, Kim, H.J, Kim, H.S. | | Deposit date: | 2023-08-22 | | Release date: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structural insights into N-terminal methionine cleavage by the human mitochondrial methionine aminopeptidase, MetAP1D.
Sci Rep, 13, 2023
|
|
8KHN
 
 | | Crystal structure of human methionine aminopeptidase 12 (MAP12) in complex with two cobalt ions | | Descriptor: | COBALT (II) ION, Methionine aminopeptidase 1D, mitochondrial, ... | | Authors: | Lee, Y, Lee, E, Hahn, H, Kim, H, Heo, Y, Jang, D.M, Kim, H.J, Kim, H.S. | | Deposit date: | 2023-08-22 | | Release date: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Structural insights into N-terminal methionine cleavage by the human mitochondrial methionine aminopeptidase, MetAP1D.
Sci Rep, 13, 2023
|
|
8KHM
 
 | | Crystal structure of human methionine aminopeptidase 12 (MAP12) in the unbound form | | Descriptor: | GLYCEROL, Methionine aminopeptidase 1D, mitochondrial, ... | | Authors: | Lee, Y, Lee, E, Hahn, H, Kim, H, Heo, Y, Jang, D.M, Kim, H.J, Kim, H.S. | | Deposit date: | 2023-08-22 | | Release date: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Structural insights into N-terminal methionine cleavage by the human mitochondrial methionine aminopeptidase, MetAP1D.
Sci Rep, 13, 2023
|
|
8WSA
 
 | | Cryo-EM Structure of Mouse TLR4/MD-2/DLAM5 Complex | | Descriptor: | (3R)-3-(dodecanoyloxy)tetradecanoic acid, (3R)-3-(tetradecanoyloxy)tetradecanoic acid, (3~{S})-3-decanoyloxytetradecanoic acid, ... | | Authors: | Fu, Y, Kim, H, Zamyatina, A, Kim, H.M. | | Deposit date: | 2023-10-17 | | Release date: | 2024-10-23 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural insight into TLR4/MD-2 activation by synthetic LPS mimetics with distinct binding modes.
Nat Commun, 16, 2025
|
|
8ZBH
 
 | | Crystal structure of TEAD3 YAP binding domain with compound 3 | | Descriptor: | 1-[7-[4-(trifluoromethyl)phenyl]-3,4-dihydro-1~{H}-isoquinolin-2-yl]propan-1-one, Transcriptional enhancer factor TEF-5 | | Authors: | Yoo, Y, Kim, H. | | Deposit date: | 2024-04-26 | | Release date: | 2025-04-16 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Pan-Transcriptional Enhanced Associated Domain Palmitoylation Pocket Covalent Inhibitor.
J.Med.Chem., 67, 2024
|
|
8ZBG
 
 | | Crystal structure of TEAD3 YAP binding domain with compound 15 | | Descriptor: | (2~{R})-2-fluoranyl-1-[7-[4-(trifluoromethyl)phenyl]-3,4-dihydro-1~{H}-isoquinolin-2-yl]propan-1-one, Transcriptional enhancer factor TEF-5 | | Authors: | Yoo, Y, Kim, H. | | Deposit date: | 2024-04-26 | | Release date: | 2025-04-16 | | Method: | X-RAY DIFFRACTION (2.67 Å) | | Cite: | Pan-Transcriptional Enhanced Associated Domain Palmitoylation Pocket Covalent Inhibitor.
J.Med.Chem., 67, 2024
|
|
9JBP
 
 | | Cryo-EM structure of human SOD1 (C6A/C111A) amyloid filament | | Descriptor: | Superoxide dismutase [Cu-Zn], Unassigned poly-alanine model | | Authors: | Baek, Y, Kim, H, Lee, D, Kim, D, Jo, E, Roh, S.-H, Ha, N.-C. | | Deposit date: | 2024-08-27 | | Release date: | 2025-01-15 | | Last modified: | 2025-02-05 | | Method: | ELECTRON MICROSCOPY (3.18 Å) | | Cite: | Structural insights into the role of reduced cysteine residues in SOD1 amyloid filament formation.
Proc.Natl.Acad.Sci.USA, 122, 2025
|
|
9JBO
 
 | | Cryo-EM structure of human SOD1 (WT) amyloid filament | | Descriptor: | Superoxide dismutase [Cu-Zn] | | Authors: | Baek, Y, Kim, H, Lee, D, Kim, D, Jo, E, Roh, S.-H, Ha, N.-C. | | Deposit date: | 2024-08-27 | | Release date: | 2025-01-15 | | Last modified: | 2025-02-05 | | Method: | ELECTRON MICROSCOPY (3.39 Å) | | Cite: | Structural insights into the role of reduced cysteine residues in SOD1 amyloid filament formation.
Proc.Natl.Acad.Sci.USA, 122, 2025
|
|
7FDE
 
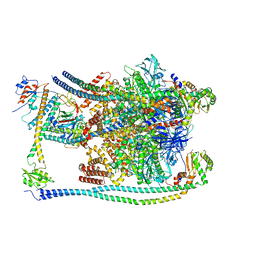 | | CryoEM Structures of Reconstituted V-ATPase, Oxr1 bound V1 | | Descriptor: | Oxidation resistance protein 1, V-type proton ATPase subunit B, V-type proton ATPase subunit C, ... | | Authors: | Khan, M.M, Lee, S, Oot, R.A, Couoh-Cardel, S, KIm, H, Wilkens, S, Roh, S.H. | | Deposit date: | 2021-07-16 | | Release date: | 2021-12-29 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | CryoEM Structures of Reconstituted V-ATPase and Oxr1-bound V1 Reveal a Novel Mechanism of Regulation.
Embo J., 2021
|
|
8WQT
 
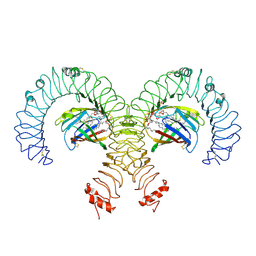 | | Cryo-EM Structure of Mouse TLR4/MD-2/DLAM1 complex | | Descriptor: | (3R)-3-(dodecanoyloxy)tetradecanoic acid, (3R)-3-(tetradecanoyloxy)tetradecanoic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Fu, Y, Kim, H, Zamyatina, A, Kim, H.M. | | Deposit date: | 2023-10-12 | | Release date: | 2024-10-16 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structural insight into TLR4/MD-2 activation by synthetic LPS mimetics with distinct binding modes.
Nat Commun, 16, 2025
|
|
8WRY
 
 | | Cryo-EM Structure of Mouse TLR4/MD-2/DLAM3 Complex | | Descriptor: | (3R)-3-(dodecanoyloxy)tetradecanoic acid, (3R)-3-(tetradecanoyloxy)tetradecanoic acid, 2-(hydroxymethyl)-5-methoxy-3,6-bis(oxidanyl)pyran-4-one, ... | | Authors: | Fu, Y, Kim, H, Zamyatina, A, Kim, H.M. | | Deposit date: | 2023-10-16 | | Release date: | 2024-10-23 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structural insight into TLR4/MD-2 activation by synthetic LPS mimetics with distinct binding modes.
Nat Commun, 16, 2025
|
|
8WTA
 
 | | Cryo-EM Structure of Human TLR4/MD-2/DLAM3 Complex | | Descriptor: | (3R)-3-(dodecanoyloxy)tetradecanoic acid, (3R)-3-(tetradecanoyloxy)tetradecanoic acid, 2-(hydroxymethyl)-5-methoxy-3,6-bis(oxidanyl)pyran-4-one, ... | | Authors: | Fu, Y, Kim, H, Zamyatina, A, Kim, H.M. | | Deposit date: | 2023-10-18 | | Release date: | 2024-10-23 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural insight into TLR4/MD-2 activation by synthetic LPS mimetics with distinct binding modes.
Nat Commun, 16, 2025
|
|
8WO1
 
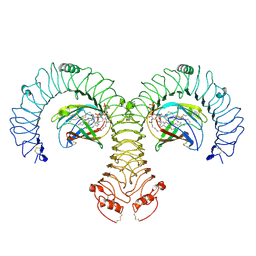 | | Cryo-EM Structure of Human TLR4/MD-2/DLAM5 Complex | | Descriptor: | (3R)-3-(dodecanoyloxy)tetradecanoic acid, (3R)-3-(tetradecanoyloxy)tetradecanoic acid, (3~{S})-3-decanoyloxytetradecanoic acid, ... | | Authors: | Fu, Y, Kim, H, Zamyatina, A, Kim, H.M. | | Deposit date: | 2023-10-06 | | Release date: | 2024-10-09 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (2.24 Å) | | Cite: | Structural insight into TLR4/MD-2 activation by synthetic LPS mimetics with distinct binding modes.
Nat Commun, 16, 2025
|
|
5X9Q
 
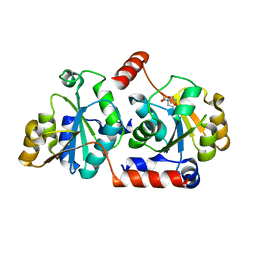 | | Crystal structure of HldC from Burkholderia pseudomallei | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Putative cytidylyltransferase | | Authors: | Park, J, Kim, H, Kim, S, Lee, D, Shin, D.H. | | Deposit date: | 2017-03-08 | | Release date: | 2017-12-27 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of D-glycero-Beta-D-manno-heptose-1-phosphate adenylyltransferase from Burkholderia pseudomallei.
Proteins, 86, 2018
|
|
6KRT
 
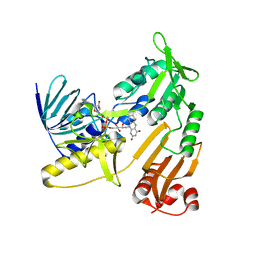 | | monodehydroascorbate reductase, MDHAR, from Antarctic hairgrass Deschampsia antarctica | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, monodehydroascorbate reductase | | Authors: | Park, A.K, Do, H, Lee, J.H, Kim, H, Choi, W, Kim, I.S, Kim, H.W. | | Deposit date: | 2019-08-22 | | Release date: | 2020-08-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | monodehydroascorbate reductase, MDHAR, from Antarctic hairgrass Deschampsia antarctica
To Be Published
|
|
5G1E
 
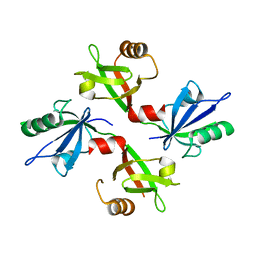 | | The complex structure of syntenin-1 PDZ domain with c-terminal extension | | Descriptor: | SYNTENIN-1 | | Authors: | Lee, I, Kim, H, Yun, J.H, Lee, W. | | Deposit date: | 2016-03-25 | | Release date: | 2016-11-23 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | New Structural Insight of C-Terminal Region of Syntenin-1, Enhancing the Molecular Dimerization and Inhibitory Function Related on Syndecan-4 Signaling.
Sci.Rep., 6, 2016
|
|
5G1D
 
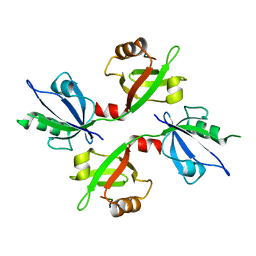 | | The complex structure of syntenin-1 PDZ domain with c-terminal extension | | Descriptor: | SYNDECAN-4, SYNTENIN-1 | | Authors: | Lee, I, Kim, H, Yun, J.H, Lee, W. | | Deposit date: | 2016-03-25 | | Release date: | 2016-11-23 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | New Structural Insight of C-Terminal Region of Syntenin-1, Enhancing the Molecular Dimerization and Inhibitory Function Related on Syndecan-4 Signaling.
Sci.Rep., 6, 2016
|
|
5XF2
 
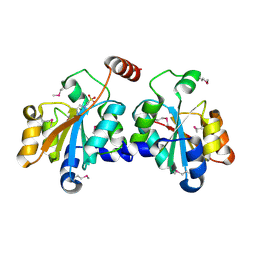 | | Crystal structure of SeMet-HldC from Burkholderia pseudomallei | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Putative cytidylyltransferase | | Authors: | Park, J, Kim, H, Kim, S, Lee, D, Shin, D.H. | | Deposit date: | 2017-04-07 | | Release date: | 2017-07-19 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Expression and crystallographic studies of D-glycero-beta-D-manno-heptose-1-phosphate adenylyltransferase from Burkholderia pseudomallei
Acta Crystallogr F Struct Biol Commun, 73, 2017
|
|
