3A4C
 
 | | Crystal structure of cdt1 C terminal domain | | Descriptor: | DNA replication factor Cdt1 | | Authors: | Cho, Y, Lee, J.H. | | Deposit date: | 2009-07-06 | | Release date: | 2009-10-13 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.889 Å) | | Cite: | Structure of the Cdt1 C-terminal domain: Conservation of the winged helix fold in replication licensing factors
Protein Sci., 18, 2009
|
|
1YLA
 
 | | Ubiquitin-conjugating enzyme E2-25 kDa (Huntington interacting protein 2) | | Descriptor: | Ubiquitin-conjugating enzyme E2-25 kDa | | Authors: | Choe, J, Avvakumov, G.V, Newman, E.M, Mackenzie, F, Kozieradzki, I, Bochkarev, A, Sundstrom, M, Arrowsmith, C, Edwards, A, Dhe-paganon, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2005-01-19 | | Release date: | 2005-02-01 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis of E2-25K/UBB+1 interaction leading to proteasome inhibition and neurotoxicity
J.Biol.Chem., 285, 2010
|
|
4LL4
 
 | | The structure of the TRX and TXNIP complex | | Descriptor: | Thioredoxin, Thioredoxin-interacting protein | | Authors: | Hwang, J, Kim, M.H. | | Deposit date: | 2013-07-09 | | Release date: | 2014-02-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The structural basis for the negative regulation of thioredoxin by thioredoxin-interacting protein
Nat Commun, 5, 2014
|
|
4LL1
 
 | | The structure of the TRX and TXNIP complex | | Descriptor: | Thioredoxin, Thioredoxin-interacting protein | | Authors: | Hwang, J, Kim, M.H. | | Deposit date: | 2013-07-09 | | Release date: | 2014-02-05 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The structural basis for the negative regulation of thioredoxin by thioredoxin-interacting protein
Nat Commun, 5, 2014
|
|
2KSE
 
 | | Backbone structure of the membrane domain of E. coli histidine kinase receptor QseC, Center for Structures of Membrane Proteins (CSMP) target 4311C | | Descriptor: | Sensor protein qseC | | Authors: | Maslennikov, I, Klammt, C, Kefala, G, Esquivies, L, Kwiatkowski, W, Choe, S, Center for Structures of Membrane Proteins (CSMP) | | Deposit date: | 2010-01-02 | | Release date: | 2010-03-02 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Membrane domain structures of three classes of histidine kinase receptors by cell-free expression and rapid NMR analysis.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3GVY
 
 | |
3K9P
 
 | | The crystal structure of E2-25K and ubiquitin complex | | Descriptor: | Ubiquitin, Ubiquitin-conjugating enzyme E2 K | | Authors: | Kang, G.B, Ko, S, Song, S.M, Lee, W, Eom, S.H. | | Deposit date: | 2009-10-16 | | Release date: | 2010-09-08 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis of E2-25K/UBB+1 interaction leading to proteasome inhibition and neurotoxicity
J.Biol.Chem., 285, 2010
|
|
5DQY
 
 | | A fully oxidized human thioredoxin | | Descriptor: | BENZOIC ACID, CHLORIDE ION, GLYCEROL, ... | | Authors: | Hwang, J. | | Deposit date: | 2015-09-15 | | Release date: | 2015-12-23 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structure of fully oxidized human thioredoxin.
Biochem.Biophys.Res.Commun., 467, 2015
|
|
2NX8
 
 | |
2KSF
 
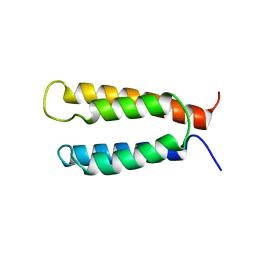 | | Backbone structure of the membrane domain of E. coli histidine kinase receptor KdpD, Center for Structures of Membrane Proteins (CSMP) target 4312C | | Descriptor: | Sensor protein kdpD | | Authors: | Maslennikov, I, Klammt, C, Kefala, G, Okamura, M, Esquivies, L, Kwiatkowski, W, Choe, S, Center for Structures of Membrane Proteins (CSMP) | | Deposit date: | 2010-01-03 | | Release date: | 2010-03-02 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Membrane domain structures of three classes of histidine kinase receptors by cell-free expression and rapid NMR analysis.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3TD5
 
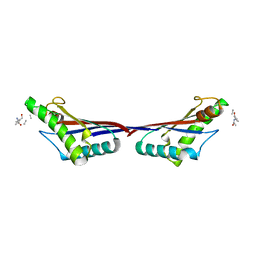 | | Crystal structure of OmpA-like domain from Acinetobacter baumannii in complex with L-Ala-gamma-D-Glu-m-DAP-D-Ala-D-Ala | | Descriptor: | CHLORIDE ION, Outer membrane protein omp38, peptide(L-Ala-gamma-D-Glu-m-DAP-D-Ala-D-Ala) | | Authors: | Park, J.S, Lee, W.C, Song, J.H, Kim, H.Y. | | Deposit date: | 2011-08-10 | | Release date: | 2011-10-26 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Mechanism of anchoring of OmpA protein to the cell wall peptidoglycan of the gram-negative bacterial outer membrane
Faseb J., 26, 2012
|
|
3TD4
 
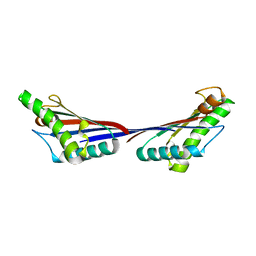 | | Crystal structure of OmpA-like domain from Acinetobacter baumannii in complex with diaminopimelate | | Descriptor: | 2,6-DIAMINOPIMELIC ACID, Outer membrane protein omp38 | | Authors: | Park, J.S, Lee, W.C, Song, J.H, Kim, H.Y. | | Deposit date: | 2011-08-10 | | Release date: | 2011-10-26 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Mechanism of anchoring of OmpA protein to the cell wall peptidoglycan of the gram-negative bacterial outer membrane
Faseb J., 26, 2012
|
|
3TD3
 
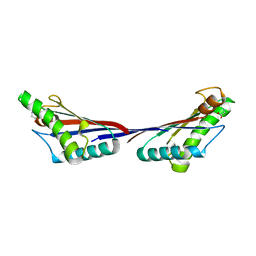 | | Crystal structure of OmpA-like domain from Acinetobacter baumannii in complex with glycine | | Descriptor: | GLYCINE, Outer membrane protein omp38 | | Authors: | Park, J.S, Lee, W.C, Song, J.H, Kim, H.Y. | | Deposit date: | 2011-08-10 | | Release date: | 2011-10-26 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Mechanism of anchoring of OmpA protein to the cell wall peptidoglycan of the gram-negative bacterial outer membrane
Faseb J., 26, 2012
|
|
3Q4R
 
 | |
3Q4N
 
 | |
3OPE
 
 | | Structural Basis of Auto-inhibitory mechanism of Histone methyltransferase | | Descriptor: | Probable histone-lysine N-methyltransferase ASH1L, S-ADENOSYLMETHIONINE, ZINC ION | | Authors: | An, S, Song, J. | | Deposit date: | 2010-08-31 | | Release date: | 2011-01-12 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of the human histone methyltransferase ASH1L catalytic domain and its implications for the regulatory mechanism
J.Biol.Chem., 286, 2011
|
|
3Q4Q
 
 | |
3Q4O
 
 | |
4K86
 
 | |
4K87
 
 | | Crystal structure of human prolyl-tRNA synthetase (substrate bound form) | | Descriptor: | ADENOSINE, PROLINE, Proline--tRNA ligase, ... | | Authors: | Hwang, K.Y, Son, J.H, Lee, E.H. | | Deposit date: | 2013-04-18 | | Release date: | 2013-10-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.301 Å) | | Cite: | Conformational changes in human prolyl-tRNA synthetase upon binding of the substrates proline and ATP and the inhibitor halofuginone.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4K88
 
 | | Crystal structure of human prolyl-tRNA synthetase (halofuginone bound form) | | Descriptor: | 7-bromo-6-chloro-3-{3-[(2R,3S)-3-hydroxypiperidin-2-yl]-2-oxopropyl}quinazolin-4(3H)-one, Proline--tRNA ligase, ZINC ION | | Authors: | Hwang, K.Y, Son, J.H, Lee, E.H. | | Deposit date: | 2013-04-18 | | Release date: | 2013-10-09 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.619 Å) | | Cite: | Conformational changes in human prolyl-tRNA synthetase upon binding of the substrates proline and ATP and the inhibitor halofuginone.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
6O76
 
 | | Human cytosolic Histidyl-tRNA synthetase (HisRS) with WHEP domain | | Descriptor: | CHLORIDE ION, Histidine--tRNA ligase, cytoplasmic | | Authors: | Kuhle, B, Yang, X.L. | | Deposit date: | 2019-03-07 | | Release date: | 2019-09-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.787 Å) | | Cite: | CMT disease severity correlates with mutation-induced open conformation of histidyl-tRNA synthetase, not aminoacylation loss, in patient cells.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6A6K
 
 | | Crystal structure of Estrogen-related Receptor-3 (ERR-gamma) ligand binding domain with DN201000 | | Descriptor: | 3-[(~{E})-5-oxidanyl-2-phenyl-1-[4-(4-propan-2-ylpiperazin-1-yl)phenyl]pent-1-enyl]phenol, Estrogen-related receptor gamma | | Authors: | Yoon, H, Kim, J, Chin, J, Cho, S.J, Song, J. | | Deposit date: | 2018-06-28 | | Release date: | 2019-04-10 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Discovery of Potent, Selective, and Orally Bioavailable Estrogen-Related Receptor-gamma Inverse Agonists To Restore the Sodium Iodide Symporter Function in Anaplastic Thyroid Cancer.
J. Med. Chem., 62, 2019
|
|
