1BCC
 
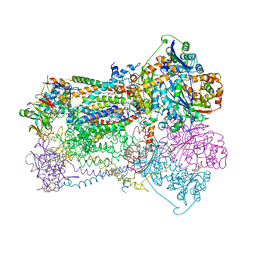 | | CYTOCHROME BC1 COMPLEX FROM CHICKEN | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, FE2/S2 (INORGANIC) CLUSTER, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Zhang, Z, Huang, L, Shulmeister, V.M, Chi, Y.-I, Kim, K.K, Hung, L.-W, Crofts, A.R, Berry, E.A, Kim, S.-H. | | Deposit date: | 1998-03-23 | | Release date: | 1998-08-19 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (3.16 Å) | | Cite: | Electron transfer by domain movement in cytochrome bc1.
Nature, 392, 1998
|
|
5MJG
 
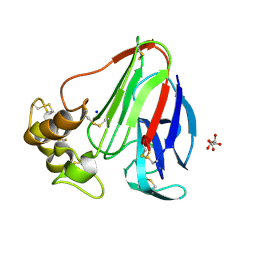 | | Single-shot pink beam serial crystallography: Thaumatin | | Descriptor: | S,R MESO-TARTARIC ACID, SODIUM ION, Thaumatin-1 | | Authors: | Meents, A, Oberthuer, D, Lieske, J, Srajer, V. | | Deposit date: | 2016-12-01 | | Release date: | 2017-12-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Single-shot pink beam serial crystallography: Thaumatin
To Be Published
|
|
7JMC
 
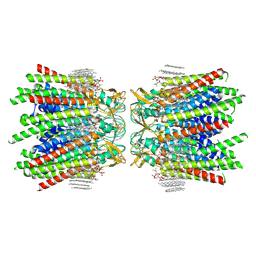 | | Sheep Connexin-50 at 2.5 angstroms resolution, Lipid Class 3 | | Descriptor: | 1,2-DIMYRISTOYL-RAC-GLYCERO-3-PHOSPHOCHOLINE, Gap junction alpha-8 protein | | Authors: | Flores, J.A, Haddad, B.G, Dolan, K.D, Myers, J.B, Yoshioka, C.C, Copperman, J, Zuckerman, D.M, Reichow, S.L. | | Deposit date: | 2020-07-31 | | Release date: | 2020-09-09 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Connexin-46/50 in a dynamic lipid environment resolved by CryoEM at 1.9 angstrom.
Nat Commun, 11, 2020
|
|
7JY2
 
 | | Z-DNA joint X-ray/Neutron | | Descriptor: | Chains: A,B | | Authors: | Harp, J.M, Coates, L, Egli, M. | | Deposit date: | 2020-08-28 | | Release date: | 2021-04-28 | | Last modified: | 2024-04-03 | | Method: | NEUTRON DIFFRACTION (1.5 Å), X-RAY DIFFRACTION | | Cite: | Water structure around a left-handed Z-DNA fragment analyzed by cryo neutron crystallography.
Nucleic Acids Res., 49, 2021
|
|
7UPH
 
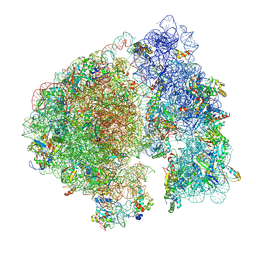 | | Structure of a ribosome with tethered subunits | | Descriptor: | 30S ribosomal protein S10, 30S ribosomal protein S11, 30S ribosomal protein S12, ... | | Authors: | Kim, D.S, Watkins, A, Bidstrup, E, Lee, J, Topkar, V.V, Kofman, C, Schwarz, K.J, Liu, Y, Pintilie, G, Roney, E, Das, R, Jewett, M.C. | | Deposit date: | 2022-04-15 | | Release date: | 2022-08-17 | | Last modified: | 2025-03-19 | | Method: | ELECTRON MICROSCOPY (4.18 Å) | | Cite: | Three-dimensional structure-guided evolution of a ribosome with tethered subunits.
Nat.Chem.Biol., 18, 2022
|
|
4Y5U
 
 | | Transcription factor | | Descriptor: | NICKEL (II) ION, Signal transducer and activator of transcription 6 | | Authors: | Li, J, Niu, F, Ouyang, S, Liu, Z. | | Deposit date: | 2015-02-12 | | Release date: | 2016-02-17 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.708 Å) | | Cite: | Structural basis for DNA recognition by STAT6
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
4Y5W
 
 | | Transcription factor-DNA complex | | Descriptor: | DNA (5'-D(P*AP*TP*GP*GP*AP*TP*TP*TP*CP*CP*TP*AP*GP*GP*AP*AP*GP*AP*CP*AP*A)-3'), DNA (5'-D(P*TP*TP*GP*TP*CP*TP*TP*CP*CP*TP*AP*GP*GP*AP*AP*AP*TP*CP*CP*AP*T)-3'), Signal transducer and activator of transcription 6 | | Authors: | Li, J, Niu, F, Ouyang, S, Liu, Z. | | Deposit date: | 2015-02-12 | | Release date: | 2016-02-17 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.104 Å) | | Cite: | Structural basis for DNA recognition by STAT6
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5MJL
 
 | | Single-shot pink beam serial crystallography: Proteinase K | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Meents, A, Oberthuer, D, Lieske, J, Srajer, V. | | Deposit date: | 2016-12-01 | | Release date: | 2017-11-15 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.21013784 Å) | | Cite: | Pink-beam serial crystallography.
Nat Commun, 8, 2017
|
|
9B9P
 
 | | Crystal structure of Staphylococcus aureus ketol-acid reductoisomerase in complex with Mg2+, and JK-5-114 | | Descriptor: | 6-hydroxy-2-phenyl[1,3]thiazolo[4,5-d]pyrimidine-5,7(4H,6H)-dione, Ketol-acid reductoisomerase (NADP(+)), MAGNESIUM ION | | Authors: | Kurz, J.L, Lin, X, Guddat, L.W. | | Deposit date: | 2024-04-03 | | Release date: | 2025-04-09 | | Last modified: | 2025-06-04 | | Method: | X-RAY DIFFRACTION (3.18 Å) | | Cite: | A ketol-acid reductoisomerase inhibitor that has antituberculosis and herbicidal activity.
Chemistry, 2025
|
|
6BX8
 
 | | Human Mesotrypsin (PRSS3) Complexed with Tissue Factor Pathway Inhibitor Variant (TFPI1-KD1-K15R-I17C-I34C) | | Descriptor: | SULFATE ION, Tissue factor pathway inhibitor, Trypsin-3 | | Authors: | Coban, M, Sankaran, B, Cohen, I, Hockla, A, Papo, N, Radisky, E.S. | | Deposit date: | 2017-12-18 | | Release date: | 2019-02-06 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Disulfide engineering of human Kunitz-type serine protease inhibitors enhances proteolytic stability and target affinity toward mesotrypsin.
J. Biol. Chem., 294, 2019
|
|
3SXM
 
 | |
3SXY
 
 | |
3SXZ
 
 | |
3SXK
 
 | |
7OS1
 
 | | Cryo-EM structure of Brr2 in complex with Fbp21 | | Descriptor: | U5 small nuclear ribonucleoprotein 200 kDa helicase, WW domain-binding protein 4 | | Authors: | Bergfort, A, Hilal, T, Weber, G, Wahl, M.C. | | Deposit date: | 2021-06-07 | | Release date: | 2022-02-23 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | The intrinsically disordered TSSC4 protein acts as a helicase inhibitor, placeholder and multi-interaction coordinator during snRNP assembly and recycling.
Nucleic Acids Res., 50, 2022
|
|
7OS2
 
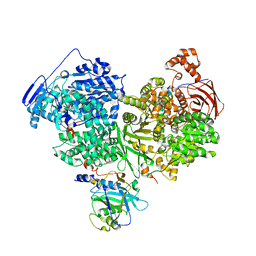 | | Cryo-EM structure of Brr2 in complex with Jab1/MPN and C9ORF78 | | Descriptor: | Pre-mRNA-processing-splicing factor 8, Telomere length and silencing protein 1 homolog, U5 small nuclear ribonucleoprotein 200 kDa helicase | | Authors: | Bergfort, A, Hilal, T, Weber, G, Wahl, M.C. | | Deposit date: | 2021-06-07 | | Release date: | 2022-02-23 | | Last modified: | 2024-09-25 | | Method: | ELECTRON MICROSCOPY (2.76 Å) | | Cite: | The intrinsically disordered TSSC4 protein acts as a helicase inhibitor, placeholder and multi-interaction coordinator during snRNP assembly and recycling.
Nucleic Acids Res., 50, 2022
|
|
5J4A
 
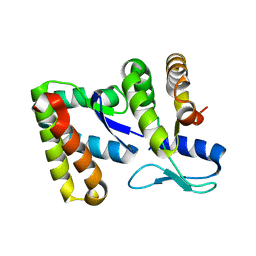 | |
4NOK
 
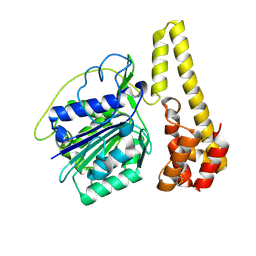 | | Crystal structure of proenzyme asparaginyl endopeptidase (AEP)/Legumain at pH 7.5 | | Descriptor: | Legumain | | Authors: | Zhao, L, Hua, T, Ru, H, Ni, X, Shaw, N, Jiao, L, Ding, W, Qu, L, Ouyang, S, Liu, Z.J. | | Deposit date: | 2013-11-19 | | Release date: | 2014-02-19 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural analysis of asparaginyl endopeptidase reveals the activation mechanism and a reversible intermediate maturation stage.
Cell Res., 24, 2014
|
|
6RPQ
 
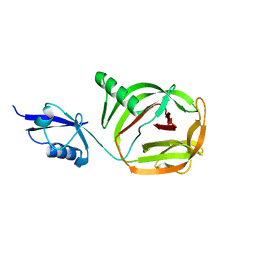 | | Crystal structure of PhoCDC21-1 intein | | Descriptor: | Ubiquitin-like protein SMT3,1108aa long hypothetical cell division control protein | | Authors: | Beyer, H.M, Mikula, K.M, Iwai, H. | | Deposit date: | 2019-05-14 | | Release date: | 2019-08-14 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.654 Å) | | Cite: | Crystal structures of CDC21-1 inteins from hyperthermophilic archaea reveal the selection mechanism for the highly conserved homing endonuclease insertion site.
Extremophiles, 23, 2019
|
|
6RPP
 
 | | Crystal structure of PabCDC21-1 intein | | Descriptor: | ACETATE ION, Cell division control protein, DI(HYDROXYETHYL)ETHER | | Authors: | Mikula, K.M, Beyer, H.M, Iwai, H. | | Deposit date: | 2019-05-14 | | Release date: | 2019-08-14 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structures of CDC21-1 inteins from hyperthermophilic archaea reveal the selection mechanism for the highly conserved homing endonuclease insertion site.
Extremophiles, 23, 2019
|
|
4NOM
 
 | | Crystal structure of asparaginyl endopeptidase (AEP)/Legumain activated at pH 4.5 | | Descriptor: | Legumain | | Authors: | Zhao, L, Hua, T, Ru, H, Ni, X, Shaw, N, Jiao, L, Ding, W, Qu, L, Ouyang, S, Liu, Z.J. | | Deposit date: | 2013-11-19 | | Release date: | 2014-02-19 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.006 Å) | | Cite: | Structural analysis of asparaginyl endopeptidase reveals the activation mechanism and a reversible intermediate maturation stage.
Cell Res., 24, 2014
|
|
6TOC
 
 | |
4PPR
 
 | | Crystal structure of Mycobacterium tuberculosis D,D-peptidase Rv3330 in complex with meropenem | | Descriptor: | (4R,5S)-3-{[(3S,5S)-5-(dimethylcarbamoyl)pyrrolidin-3-yl]sulfanyl}-5-[(2S,3R)-3-hydroxy-1-oxobutan-2-yl]-4-methyl-4,5-d ihydro-1H-pyrrole-2-carboxylic acid, Penicillin-binding protein DacB1 | | Authors: | Prigozhin, D.M, Huizar, J.P, Mavrici, D, Alber, T, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2014-02-27 | | Release date: | 2014-11-05 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Subfamily-specific adaptations in the structures of two penicillin-binding proteins from Mycobacterium tuberculosis.
Plos One, 9, 2014
|
|
4NOJ
 
 | | Crystal structure of the mature form of asparaginyl endopeptidase (AEP)/Legumain activated at pH 3.5 | | Descriptor: | Legumain | | Authors: | Zhao, L, Hua, T, Ru, H, Ni, X, Shaw, N, Jiao, L, Ding, W, Qu, L, Ouyang, S, Liu, Z.J. | | Deposit date: | 2013-11-19 | | Release date: | 2014-02-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural analysis of asparaginyl endopeptidase reveals the activation mechanism and a reversible intermediate maturation stage.
Cell Res., 24, 2014
|
|
4NOL
 
 | | Crystal structure of proenzyme asparaginyl endopeptidase (AEP)/Legumain mutant D233A at pH 7.5 | | Descriptor: | Legumain | | Authors: | Zhao, L, Hua, T, Ru, H, Ni, X, Shaw, N, Jiao, L, Ding, W, Qu, L, Ouyang, S, Liu, Z.J. | | Deposit date: | 2013-11-19 | | Release date: | 2014-02-19 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural analysis of asparaginyl endopeptidase reveals the activation mechanism and a reversible intermediate maturation stage.
Cell Res., 24, 2014
|
|
