6KIU
 
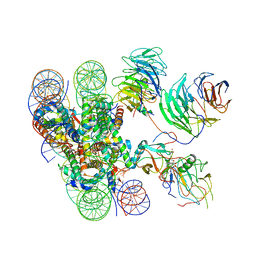 | | Cryo-EM structure of human MLL1-ubNCP complex (3.2 angstrom) | | Descriptor: | DNA (145-MER), GLUTAMINE, Histone H2A, ... | | Authors: | Huang, J, Xue, H, Yao, T. | | Deposit date: | 2019-07-20 | | Release date: | 2019-09-11 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural basis of nucleosome recognition and modification by MLL methyltransferases.
Nature, 573, 2019
|
|
6KIW
 
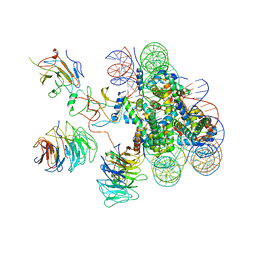 | | Cryo-EM structure of human MLL3-ubNCP complex (4.0 angstrom) | | Descriptor: | DNA (144-MER), DNA (145-MER), Histone H2A, ... | | Authors: | Huang, J, Xue, H, Yao, T. | | Deposit date: | 2019-07-20 | | Release date: | 2019-09-11 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structural basis of nucleosome recognition and modification by MLL methyltransferases.
Nature, 573, 2019
|
|
5BPP
 
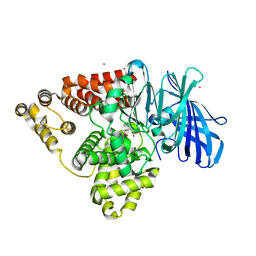 | | Structure of human Leukotriene A4 hydrolase in complex with inhibitor 4AZ | | Descriptor: | 2-(4-butoxyphenyl)-N-hydroxyacetamide, ACETATE ION, Leukotriene A-4 hydrolase, ... | | Authors: | Huang, J, Dong, N.N, Xiao, Q, Ou, P.Y, Wu, D, Lu, W.Q. | | Deposit date: | 2015-05-28 | | Release date: | 2016-08-10 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Bufexamac ameliorates LPS-induced acute lung injury in mice by targeting LTA4H
Sci Rep, 6, 2016
|
|
4XX0
 
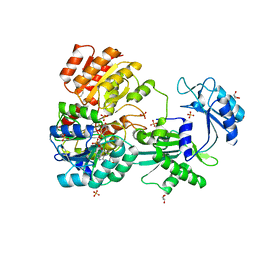 | | CoA bound to pig GTP-specific succinyl-CoA synthetase | | Descriptor: | COENZYME A, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Fraser, M.E, Huang, J, Malhi, M. | | Deposit date: | 2015-01-29 | | Release date: | 2015-08-12 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of GTP-specific succinyl-CoA synthetase in complex with CoA.
Acta Crystallogr.,Sect.F, 71, 2015
|
|
8FBC
 
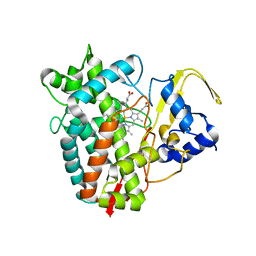 | | Crystal structure of P450T2 | | Descriptor: | Cytochrome P450, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Pereira, J.H, Huang, J, Keasling, J, Adams, P.D. | | Deposit date: | 2022-11-29 | | Release date: | 2023-05-10 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Complete integration of carbene-transfer chemistry into biosynthesis.
Nature, 617, 2023
|
|
8G1A
 
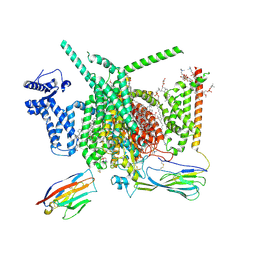 | | Cryo-EM structure of Nav1.7 with CBD | | Descriptor: | (3beta,14beta,17beta,25R)-3-[4-methoxy-3-(methoxymethyl)butoxy]spirost-5-en, 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1-O-OCTADECYL-SN-GLYCERO-3-PHOSPHOCHOLINE, ... | | Authors: | Fan, X, Huang, J, Yan, N. | | Deposit date: | 2023-02-01 | | Release date: | 2023-07-05 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Cannabidiol inhibits Na v channels through two distinct binding sites.
Nat Commun, 14, 2023
|
|
5HC0
 
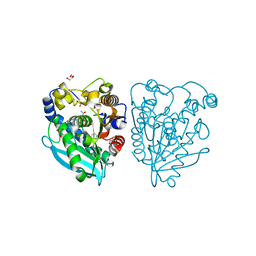 | | Structure of esterase Est22 with p-nitrophenol | | Descriptor: | ACETIC ACID, GLYCEROL, Lipolytic enzyme, ... | | Authors: | Li, J, Huang, J. | | Deposit date: | 2016-01-04 | | Release date: | 2017-01-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural insights of a hormone sensitive lipase homologue Est22.
Sci Rep, 6, 2016
|
|
5HC5
 
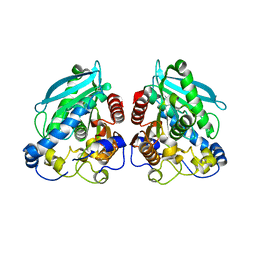 | |
5HC2
 
 | |
5HC3
 
 | | The structure of esterase Est22 | | Descriptor: | GLYCEROL, Lipolytic enzyme | | Authors: | Li, J, Huang, J. | | Deposit date: | 2016-01-04 | | Release date: | 2017-01-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural insights of a hormone sensitive lipase homologue Est22.
Sci Rep, 6, 2016
|
|
8SX3
 
 | | 10E8-GT10.2 immunogen in complex with human Fab 10E8 and mouse Fab W6-10 | | Descriptor: | 10E8 Fab heavy chain, 10E8 light chain, 10E8-GT10.2 immunogen, ... | | Authors: | Huang, J, Ozorowski, G, Ward, A.B. | | Deposit date: | 2023-05-19 | | Release date: | 2024-05-22 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Vaccination induces broadly neutralizing antibody precursors to HIV gp41.
Nat.Immunol., 25, 2024
|
|
5HC4
 
 | | Structure of esterase Est22 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GLYCEROL, Lipolytic enzyme | | Authors: | Li, J, Huang, J. | | Deposit date: | 2016-01-04 | | Release date: | 2017-01-18 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural insights of a hormone sensitive lipase homologue Est22.
Sci Rep, 6, 2016
|
|
8UT7
 
 | | CryoEM structure of A/Perth/16/2009 H3 in complex with polyclonal Fab from mice immunized with H3 stem nanoparticles-28 days post immunization | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, H3D28 pFab HC Fv_polyA, ... | | Authors: | Huang, J, Han, J, Ward, A.B. | | Deposit date: | 2023-10-30 | | Release date: | 2024-05-08 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Eliciting a single amino acid change by vaccination generates antibody protection against group 1 and group 2 influenza A viruses.
Immunity, 57, 2024
|
|
8UT6
 
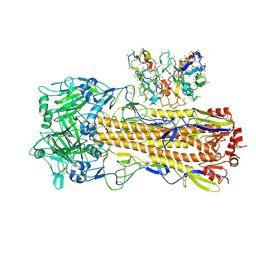 | | CryoEM structure of A/Perth/16/2009 H3 in complex with polyclonal Fab from mice immunized with H3 stem nanoparticles-15 days post immunization | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, H3D15 pFab HC Fv_polyA, ... | | Authors: | Huang, J, Han, J, Ward, A.B. | | Deposit date: | 2023-10-30 | | Release date: | 2024-05-08 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Eliciting a single amino acid change by vaccination generates antibody protection against group 1 and group 2 influenza A viruses.
Immunity, 57, 2024
|
|
8UT9
 
 | | CryoEM structure of A/Shanghai/1/2013 H7 in complex with polyclonal Fab from mice immunized with H7 stem nanoparticles-28 days post immunization | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Huang, J, Han, J, Ward, A.B. | | Deposit date: | 2023-10-30 | | Release date: | 2024-05-08 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Eliciting a single amino acid change by vaccination generates antibody protection against group 1 and group 2 influenza A viruses.
Immunity, 57, 2024
|
|
8UT3
 
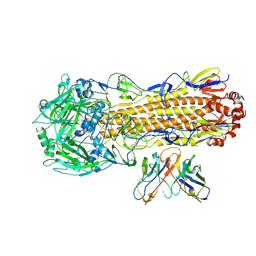 | | CryoEM structure of A/Perth/16/2009 H3 in complex with flu HA central stem VH1-18 antibody UCA6 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin, ... | | Authors: | Huang, J, Han, J, Ward, A.B. | | Deposit date: | 2023-10-30 | | Release date: | 2024-05-08 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Eliciting a single amino acid change by vaccination generates antibody protection against group 1 and group 2 influenza A viruses.
Immunity, 57, 2024
|
|
8UT8
 
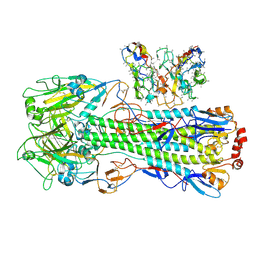 | | CryoEM structure of A/Shanghai/1/2013 H7 in complex with polyclonal Fab from mice immunized with H7 stem nanoparticles-15 days post-immunization | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Huang, J, Han, J, Ward, A.B. | | Deposit date: | 2023-10-30 | | Release date: | 2024-05-08 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Eliciting a single amino acid change by vaccination generates antibody protection against group 1 and group 2 influenza A viruses.
Immunity, 57, 2024
|
|
8UT4
 
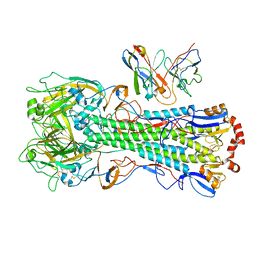 | | CryoEM structure of A/Michigan/45/2015 H1 in complex with flu HA central stem VH1-18 antibody 09-1B12 | | Descriptor: | 09-1B12 HC Fv, 09-1B12 LC Fv, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Huang, J, Han, J, Ward, A.B. | | Deposit date: | 2023-10-30 | | Release date: | 2024-05-08 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Eliciting a single amino acid change by vaccination generates antibody protection against group 1 and group 2 influenza A viruses.
Immunity, 57, 2024
|
|
8UT5
 
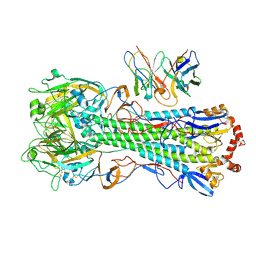 | | CryoEM structure of A/Michigan/45/2015 H1 in complex with flu HA central stem VH1-18 antibody UCA6_N55T | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin HA1 chain, Hemagglutinin HA2 chain, ... | | Authors: | Huang, J, Han, J, Ward, A.B. | | Deposit date: | 2023-10-30 | | Release date: | 2024-05-08 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Eliciting a single amino acid change by vaccination generates antibody protection against group 1 and group 2 influenza A viruses.
Immunity, 57, 2024
|
|
5C13
 
 | |
8T1R
 
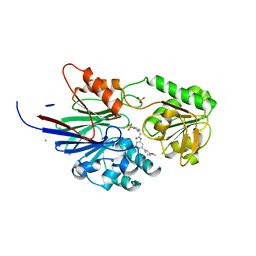 | | Crystal structure of human CPSF73 catalytic segment in complex with compound 2 | | Descriptor: | 3-[7,7-bis(oxidanyl)-8-oxa-7-boranuidabicyclo[4.3.0]nona-1,3,5-trien-5-yl]-~{N}-[3-(3-methoxyphenyl)phenyl]propanamide, CHLORIDE ION, Cleavage and polyadenylation specificity factor subunit 3, ... | | Authors: | Huang, J, Tong, L. | | Deposit date: | 2023-06-02 | | Release date: | 2023-11-08 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Anticancer benzoxaboroles block pre-mRNA processing by directly inhibiting CPSF3.
Cell Chem Biol, 31, 2024
|
|
8T1Q
 
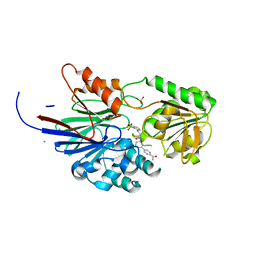 | | Crystal structure of human CPSF73 catalytic segment in complex with compound 1 | | Descriptor: | 3-[7,7-bis(oxidanyl)-8-oxa-7-boranuidabicyclo[4.3.0]nona-1,3,5-trien-5-yl]-~{N}-[3-(4-ethanoylphenyl)phenyl]propanamide, CHLORIDE ION, Cleavage and polyadenylation specificity factor subunit 3, ... | | Authors: | Huang, J, Tong, L. | | Deposit date: | 2023-06-02 | | Release date: | 2023-11-08 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Anticancer benzoxaboroles block pre-mRNA processing by directly inhibiting CPSF3.
Cell Chem Biol, 31, 2024
|
|
8WKT
 
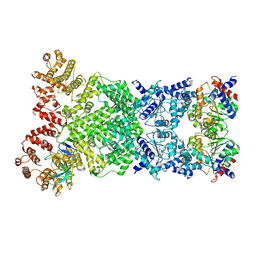 | | Cryo-EM structure of DSR2-DSAD1 complex | | Descriptor: | SIR2-like domain-containing protein, SPbeta prophage-derived uncharacterized protein YotI | | Authors: | Gao, A, Huang, J, Zhu, K. | | Deposit date: | 2023-09-28 | | Release date: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.86 Å) | | Cite: | Molecular basis of bacterial DSR2 anti-phage defense and viral immune evasion.
Nat Commun, 15, 2024
|
|
8WKX
 
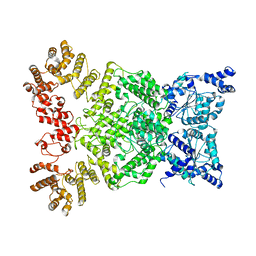 | | Cryo-EM structure of DSR2 | | Descriptor: | SIR2-like domain-containing protein | | Authors: | Gao, A, Huang, J, Zhu, K. | | Deposit date: | 2023-09-28 | | Release date: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (4.15 Å) | | Cite: | Molecular basis of bacterial DSR2 anti-phage defense and viral immune evasion.
Nat Commun, 15, 2024
|
|
8WKS
 
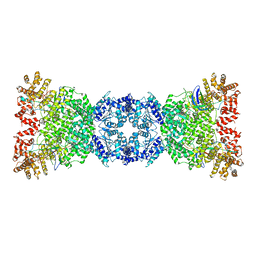 | | Cryo-EM structure of DSR2-TUBE complex | | Descriptor: | SIR2-like domain-containing protein, TUBE | | Authors: | Gao, A, Huang, J, Zhu, K. | | Deposit date: | 2023-09-28 | | Release date: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.58 Å) | | Cite: | Molecular basis of bacterial DSR2 anti-phage defense and viral immune evasion.
Nat Commun, 15, 2024
|
|
