2Z5C
 
 | | Crystal Structure of a Novel Chaperone Complex for Yeast 20S Proteasome Assembly | | Descriptor: | Proteasome component PUP2, Protein YPL144W, Uncharacterized protein YLR021W | | Authors: | Yashiroda, H, Mizushima, T, Okamoto, K, Kameyama, T, Hayashi, H, Kishimoto, T, Kasahara, M, Kurimoto, E, Sakata, E, Suzuki, A, Hirano, Y, Murata, S, Kato, K, Yamane, T, Tanaka, K. | | Deposit date: | 2007-07-03 | | Release date: | 2008-01-22 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of a chaperone complex that contributes to the assembly of yeast 20S proteasomes
Nat.Struct.Mol.Biol., 15, 2008
|
|
2Z5E
 
 | | Crystal Structure of Proteasome Assembling Chaperone 3 | | Descriptor: | Proteasome Assembling Chaperone 3 | | Authors: | Okamoto, K, Kurimoto, E, Sakata, E, Suzuki, A, Yamane, T, Hirano, Y, Murata, S, Tanaka, K, Kato, K. | | Deposit date: | 2007-07-06 | | Release date: | 2008-02-19 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of a chaperone complex that contributes to the assembly of yeast 20S proteasomes
Nat.Struct.Mol.Biol., 15, 2008
|
|
2ZSI
 
 | | Structural basis of gibberellin(GA4)-induced DELLA recognition by the gibberellin receptor | | Descriptor: | DELLA protein GAI, GIBBERELLIN A4, Probable gibberellin receptor GID1L1 | | Authors: | Murase, K, Hirano, Y, Sun, T.P, Hakoshima, T. | | Deposit date: | 2008-09-10 | | Release date: | 2008-11-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Gibberellin-induced DELLA recognition by the gibberellin receptor GID1
Nature, 456, 2008
|
|
3WX1
 
 | | Mouse Cereblon thalidomide binding domain, selenomethionine derivative | | Descriptor: | Protein cereblon, SULFATE ION, ZINC ION | | Authors: | Mori, T, Ito, T, Hirano, Y, Yamaguchi, Y, Handa, H, Hakoshima, T. | | Deposit date: | 2014-07-10 | | Release date: | 2014-08-06 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Structure of the human Cereblon-DDB1-lenalidomide complex reveals basis for responsiveness to thalidomide analogs
Nat.Struct.Mol.Biol., 21, 2014
|
|
3WX2
 
 | | Mouse Cereblon thalidomide binding domain, native | | Descriptor: | Protein cereblon, SULFATE ION, ZINC ION | | Authors: | Mori, T, Ito, T, Hirano, Y, Yamaguchi, Y, Handa, H, Hakoshima, T. | | Deposit date: | 2014-07-10 | | Release date: | 2014-08-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the human Cereblon-DDB1-lenalidomide complex reveals basis for responsiveness to thalidomide analogs
Nat.Struct.Mol.Biol., 21, 2014
|
|
3W91
 
 | | crystal structure of SeMet-labeled yeast N-acetyltransferase Mpr1 L87M mutant | | Descriptor: | MPR1 protein | | Authors: | Nasuno, R, Hirano, Y, Itoh, T, Hakoshima, T, Hibi, T, Takagi, H. | | Deposit date: | 2013-03-23 | | Release date: | 2013-07-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural and functional analysis of the yeast N-acetyltransferase Mpr1 involved in oxidative stress tolerance via proline metabolism
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
3W6X
 
 | | Yeast N-acetyltransferase Mpr1 in complex with CHOP | | Descriptor: | (4S)-4-hydroxy-L-proline, CHLORIDE ION, HEXAETHYLENE GLYCOL, ... | | Authors: | Nasuno, R, Hirano, Y, Itoh, T, Hakoshima, T, Hibi, T, Takagi, H. | | Deposit date: | 2013-02-25 | | Release date: | 2013-08-07 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.299 Å) | | Cite: | Structural and functional analysis of the yeast N-acetyltransferase Mpr1 involved in oxidative stress tolerance via proline metabolism
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
3W9R
 
 | | Crystal structure of the high-affinity abscisic acid receptor PYL9/RCAR9 bound to ABA | | Descriptor: | (2Z,4E)-5-[(1S)-1-hydroxy-2,6,6-trimethyl-4-oxocyclohex-2-en-1-yl]-3-methylpenta-2,4-dienoic acid, Abscisic acid receptor PYL9, HEXAETHYLENE GLYCOL | | Authors: | Nakagawa, M, Hirano, Y, Kagiyama, M, Shibata, N, Hakoshima, T. | | Deposit date: | 2013-04-13 | | Release date: | 2014-04-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Mechanism of high-affinity abscisic acid binding to PYL9/RCAR1.
Genes Cells, 19, 2014
|
|
3W6S
 
 | | yeast N-acetyltransferase Mpr1 involved in oxidative stress tolerance via proline metabolism | | Descriptor: | HEXAETHYLENE GLYCOL, MAGNESIUM ION, MPR1 protein | | Authors: | Nasuno, R, Hirano, Y, Itoh, T, Hakoshima, T, Hibi, T, Takagi, H. | | Deposit date: | 2013-02-21 | | Release date: | 2013-07-17 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and functional analysis of the yeast N-acetyltransferase Mpr1 involved in oxidative stress tolerance via proline metabolism
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4V8K
 
 | | Crystal structure of the LH1-RC complex from Thermochromatium tepidum in P21 form | | Descriptor: | BACTERIOCHLOROPHYLL A, BACTERIOPHEOPHYTIN A, CALCIUM ION, ... | | Authors: | Niwa, S, Takeda, K, Wang-Otomo, Z.-Y, Miki, K. | | Deposit date: | 2013-11-22 | | Release date: | 2014-07-09 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.006 Å) | | Cite: | Structure of the LH1-RC complex from Thermochromatium tepidum at 3.0 angstrom
Nature, 508, 2014
|
|
4TZC
 
 | | Crystal Structure of Murine Cereblon in Complex with Thalidomide | | Descriptor: | Protein cereblon, S-Thalidomide, SULFATE ION, ... | | Authors: | Chamberlain, P.P, Pagarigan, B, Delker, S, Leon, B. | | Deposit date: | 2014-07-10 | | Release date: | 2014-08-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Structural Basis for Responsiveness to Thalidomide-Analog Drugs Defined by the Crystal Structure of the Human Cereblon:DDB1:Lenalidomide Complex
to be published
|
|
7XVX
 
 | |
6KYW
 
 | | S8-mSRK-S8-SP11 complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Receptor protein kinase SRK8, S locus protein 11 | | Authors: | Murase, K, Hakoshima, T, Mori, T. | | Deposit date: | 2019-09-20 | | Release date: | 2020-09-16 | | Last modified: | 2020-10-21 | | Method: | X-RAY DIFFRACTION (2.60112739 Å) | | Cite: | Mechanism of self/nonself-discrimination in Brassica self-incompatibility.
Nat Commun, 11, 2020
|
|
8W6X
 
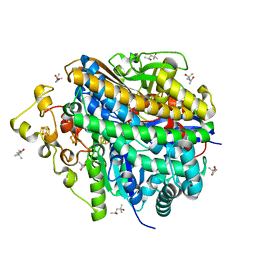 | | Neutron structure of [NiFe]-hydrogenase from D. vulgaris Miyazaki F in its oxidized state | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CHLORIDE ION, FE3-S4 CLUSTER, ... | | Authors: | Hiromoto, T, Tamada, T. | | Deposit date: | 2023-08-30 | | Release date: | 2023-09-13 | | Last modified: | 2023-11-15 | | Method: | NEUTRON DIFFRACTION (1.04 Å), X-RAY DIFFRACTION | | Cite: | New insights into the oxidation process from neutron and X-ray crystal structures of an O 2 -sensitive [NiFe]-hydrogenase.
Chem Sci, 14, 2023
|
|
7CQF
 
 | |
5ZEQ
 
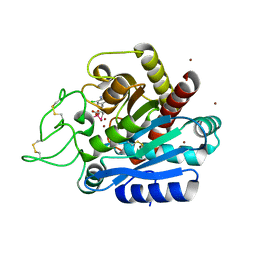 | | Carboxypeptidase B in complex with DD28 | | Descriptor: | (2~{S})-2-[(6-azanyl-5-chloranyl-pyridin-3-yl)methyl]-3-selanyl-propanoic acid, CACODYLATE ION, Carboxypeptidase B, ... | | Authors: | Itoh, T, Yoshimoto, N, Yamamoto, K. | | Deposit date: | 2018-02-27 | | Release date: | 2018-06-20 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for the selective inhibition of activated thrombin-activatable fibrinolysis inhibitor (TAFIa) by a selenium-containing inhibitor with chloro-aminopyridine as a basic group
Bioorg. Med. Chem. Lett., 28, 2018
|
|
3WMM
 
 | | Crystal structure of the LH1-RC complex from Thermochromatium tepidum in C2 form | | Descriptor: | (1R)-2-{[(S)-{[(2S)-2,3-dihydroxypropyl]oxy}(hydroxy)phosphoryl]oxy}-1-[(hexadecanoyloxy)methyl]ethyl (9Z)-octadec-9-enoate, BACTERIOCHLOROPHYLL A, BACTERIOPHEOPHYTIN A, ... | | Authors: | Niwa, S, Takeda, K, Wang-Otomo, Z.-Y, Miki, K. | | Deposit date: | 2013-11-22 | | Release date: | 2014-04-09 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.008 Å) | | Cite: | Structure of the LH1-RC complex from Thermochromatium tepidum at 3.0 angstrom
Nature, 508, 2014
|
|
