7RM8
 
 | |
5YK9
 
 | |
5Z43
 
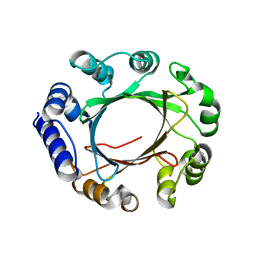 | | Crystal structure of prenyltransferase AmbP1 apo structure | | Descriptor: | AmbP1, MAGNESIUM ION | | Authors: | Awakawa, T, Nakashima, Y, Mori, T, Abe, I. | | Deposit date: | 2018-01-10 | | Release date: | 2018-06-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.361 Å) | | Cite: | Molecular Insight into the Mg2+-Dependent Allosteric Control of Indole Prenylation by Aromatic Prenyltransferase AmbP1
Angew. Chem. Int. Ed. Engl., 57, 2018
|
|
5Z45
 
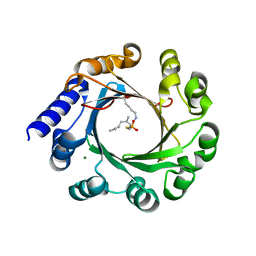 | | Crystal structure of prenyltransferase AmbP1 pH6.5 complexed with GSPP and cis-indolyl vinyl isonitrile | | Descriptor: | 3-[(Z)-2-isocyanoethenyl]-1H-indole, AmbP1, GERANYL S-THIOLODIPHOSPHATE, ... | | Authors: | Awakawa, T, Nakashima, Y, Mori, T, Abe, I. | | Deposit date: | 2018-01-10 | | Release date: | 2018-06-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.601 Å) | | Cite: | Molecular Insight into the Mg2+-Dependent Allosteric Control of Indole Prenylation by Aromatic Prenyltransferase AmbP1
Angew. Chem. Int. Ed. Engl., 57, 2018
|
|
5Z44
 
 | | Crystal structure of prenyltransferase AmbP1 complexed with GSPP | | Descriptor: | AmbP1, GERANYL S-THIOLODIPHOSPHATE, MAGNESIUM ION | | Authors: | Awakawa, T, Nakashima, Y, Mori, T, Abe, I. | | Deposit date: | 2018-01-10 | | Release date: | 2018-06-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.458 Å) | | Cite: | Molecular Insight into the Mg2+-Dependent Allosteric Control of Indole Prenylation by Aromatic Prenyltransferase AmbP1
Angew. Chem. Int. Ed. Engl., 57, 2018
|
|
5Z46
 
 | | Crystal structure of prenyltransferase AmbP1 pH8 complexed with GSPP and cis-indolyl vinyl isonitrile | | Descriptor: | 3-[(Z)-2-isocyanoethenyl]-1H-indole, AmbP1, GERANYL S-THIOLODIPHOSPHATE, ... | | Authors: | Awakawa, T, Nakashima, Y, Mori, T, Abe, I. | | Deposit date: | 2018-01-10 | | Release date: | 2018-06-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.999 Å) | | Cite: | Molecular Insight into the Mg2+-Dependent Allosteric Control of Indole Prenylation by Aromatic Prenyltransferase AmbP1
Angew. Chem. Int. Ed. Engl., 57, 2018
|
|
4RQI
 
 | | Structure of TRF2/RAP1 secondary interaction binding site | | Descriptor: | GLYCEROL, MAGNESIUM ION, Telomeric repeat-binding factor 2, ... | | Authors: | Miron, S, Guimaraes, B, Gaullier, G, Giraud-Panis, M.-J, Gilson, E, Le Du, M.-H. | | Deposit date: | 2014-11-03 | | Release date: | 2016-02-10 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.4405 Å) | | Cite: | A higher-order entity formed by the flexible assembly of RAP1 with TRF2.
Nucleic Acids Res., 44, 2016
|
|
6H8H
 
 | |
6H8B
 
 | | Molybdenum storage protein prepared under in vivo-like conditions and incubated with ATP and molybdate at 303 K | | Descriptor: | 2,2,4-tris(oxidanyl)-1,3-dioxa-2$l^{4},4$l^{3}-dimolybdacyclobutane, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Ermler, U, Poppe, J. | | Deposit date: | 2018-08-02 | | Release date: | 2019-02-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The molybdenum storage protein - A bionanolab for creating experimentally alterable polyoxomolybdate clusters.
J. Inorg. Biochem., 189, 2018
|
|
3G5F
 
 | | Crystallographic analysis of cytochrome P450 cyp121 | | Descriptor: | Cytochrome P450 121, PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION | | Authors: | Belin, P, Le Du, M.H, Gondry, M. | | Deposit date: | 2009-02-05 | | Release date: | 2009-04-21 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Identification and structural basis of the reaction catalyzed by CYP121, an essential cytochrome P450 in Mycobacterium tuberculosis.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3G5H
 
 | | Crystallographic analysis of cytochrome P450 cyp121 | | Descriptor: | (3S,6S)-3,6-bis(4-hydroxybenzyl)piperazine-2,5-dione, Cytochrome P450 121, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Belin, P, Le Du, M.H, Gondry, M. | | Deposit date: | 2009-02-05 | | Release date: | 2009-04-21 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Identification and structural basis of the reaction catalyzed by CYP121, an essential cytochrome P450 in Mycobacterium tuberculosis.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
2JWZ
 
 | | Mutations in the hydrophobic core of ubiquitin differentially affect its recognition by receptor proteins | | Descriptor: | Ubiquitin | | Authors: | Haririnia, A, Verma, R, Purohit, N, Twarog, M, Deshaies, R, Bolon, D, Fushman, D. | | Deposit date: | 2007-10-31 | | Release date: | 2008-01-08 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Mutations in the hydrophobic core of ubiquitin differentially affect its recognition by receptor proteins.
J.Mol.Biol., 375, 2008
|
|
1HAR
 
 | |
7LMS
 
 | |
7ENB
 
 | |
6ZT0
 
 | | Crystal structure of the Eiger TNF domain/Grindelwald extracellular domain complex | | Descriptor: | 1,2-ETHANEDIOL, Protein eiger, Protein grindelwald | | Authors: | Palmerini, V, Cecatiello, V, Pasqualato, S, Mapelli, M. | | Deposit date: | 2020-07-17 | | Release date: | 2021-03-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Drosophila TNFRs Grindelwald and Wengen bind Eiger with different affinities and promote distinct cellular functions.
Nat Commun, 12, 2021
|
|
7EMZ
 
 | |
7ABM
 
 | | X-ray structure of phosphorylated Barrier-to-autointegration factor (BAF) | | Descriptor: | Barrier-to-autointegration factor, CESIUM ION | | Authors: | Zinn-Justin, S, Marcelot, A, Le Du, M.H, Ropars, V. | | Deposit date: | 2020-09-08 | | Release date: | 2021-07-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.004 Å) | | Cite: | Di-phosphorylated BAF shows altered structural dynamics and binding to DNA, but interacts with its nuclear envelope partners.
Nucleic Acids Res., 49, 2021
|
|
6EYU
 
 | | Crystal structure of the inward H(+) pump xenorhodopsin | | Descriptor: | Bacteriorhodopsin, EICOSANE, RETINAL, ... | | Authors: | Kovalev, K, Shevchenko, V, Polovinkin, V, Mager, T, Gushchin, I, Melnikov, I, Borshchevskiy, V, Popov, A, Alekseev, A, Gordeliy, V. | | Deposit date: | 2017-11-13 | | Release date: | 2017-12-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Inward H(+) pump xenorhodopsin: Mechanism and alternative optogenetic approach.
Sci Adv, 3, 2017
|
|
6GUJ
 
 | | Molybdenum storage protein with two occupied ATP binding sites | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, MOLYBDATE ION, ... | | Authors: | Ermler, U, Poppe, J, Bruenle, S. | | Deposit date: | 2018-06-19 | | Release date: | 2018-11-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.102 Å) | | Cite: | The Molybdenum Storage Protein: A soluble ATP hydrolysis-dependent molybdate pump.
FEBS J., 285, 2018
|
|
6GU5
 
 | | Mosto containing the core POM clusters | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MOLYBDATE ION, Molybdenum storage protein subunit alpha, ... | | Authors: | Ermler, U, Poppe, J, Bruenle, S. | | Deposit date: | 2018-06-19 | | Release date: | 2018-11-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Molybdenum Storage Protein: A soluble ATP hydrolysis-dependent molybdate pump.
FEBS J., 285, 2018
|
|
6GWB
 
 | | Molybdenum storage protein without polyoxomolybdate clusters | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Ermler, U. | | Deposit date: | 2018-06-22 | | Release date: | 2018-10-24 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The molybdenum storage protein - A bionanolab for creating experimentally alterable polyoxomolybdate clusters.
J. Inorg. Biochem., 189, 2018
|
|
6H74
 
 | | The molybdenum storage protein - L131H | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, MOLYBDATE ION, ... | | Authors: | Ermler, U, Bruenle, S. | | Deposit date: | 2018-07-30 | | Release date: | 2018-11-14 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | The molybdenum storage protein - A bionanolab for creating experimentally alterable polyoxomolybdate clusters.
J. Inorg. Biochem., 189, 2018
|
|
6H73
 
 | |
6GWV
 
 | | Molybdenum storage protein without polymolybdate clusters and ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CHLORIDE ION, Molybdenum storage protein subunit alpha, ... | | Authors: | Ermler, U, Poppe, J, Bruenle, S. | | Deposit date: | 2018-06-26 | | Release date: | 2018-11-07 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The Molybdenum Storage Protein: A soluble ATP hydrolysis-dependent molybdate pump.
FEBS J., 285, 2018
|
|
