4BOQ
 
 | | Structure of OTUD2 OTU domain | | Descriptor: | GLYCEROL, UBIQUITIN THIOESTERASE OTU1 | | Authors: | Mevissen, T.E.T, Hospenthal, M.K, Geurink, P.P, Elliott, P.R, Akutsu, M, Arnaudo, N, Ekkebus, R, Kulathu, Y, Wauer, T, El Oualid, F, Freund, S.M.V, Ovaa, H, Komander, D. | | Deposit date: | 2013-05-22 | | Release date: | 2013-07-17 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Otu Deubiquitinases Reveal Mechanisms of Linkage Specificity and Enable Ubiquitin Chain Restriction Analysis.
Cell(Cambridge,Mass.), 154, 2013
|
|
4BOU
 
 | | Structure of OTUD3 OTU domain | | Descriptor: | OTU DOMAIN-CONTAINING PROTEIN 3 | | Authors: | Mevissen, T.E.T, Hospenthal, M.K, Geurink, P.P, Elliott, P.R, Akutsu, M, Arnaudo, N, Ekkebus, R, Kulathu, Y, Wauer, T, El Oualid, F, Freund, S.M.V, Ovaa, H, Komander, D. | | Deposit date: | 2013-05-22 | | Release date: | 2013-07-17 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Otu Deubiquitinases Reveal Mechanisms of Linkage Specificity and Enable Ubiquitin Chain Restriction Analysis.
Cell(Cambridge,Mass.), 154, 2013
|
|
4BOS
 
 | | Structure of OTUD2 OTU domain in complex with Ubiquitin K11-linked peptide | | Descriptor: | MAGNESIUM ION, NITRATE ION, OTUD2, ... | | Authors: | Mevissen, T.E.T, Hospenthal, M.K, Geurink, P.P, Elliott, P.R, Akutsu, M, Arnaudo, N, Ekkebus, R, Kulathu, Y, Wauer, T, El Oualid, F, Freund, S.M.V, Ovaa, H, Komander, D. | | Deposit date: | 2013-05-22 | | Release date: | 2013-07-24 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Otu Deubiquitinases Reveal Mechanisms of Linkage Specificity and Enable Ubiquitin Chain Restriction Analysis.
Cell(Cambridge,Mass.), 154, 2013
|
|
4CH2
 
 | | Low-salt crystal structure of a thrombin-GpIbalpha peptide complex | | Descriptor: | D-phenylalanyl-N-[(2S,3S)-6-{[amino(iminio)methyl]amino}-1-chloro-2-hydroxyhexan-3-yl]-L-prolinamide, GLYCEROL, PLATELET GLYCOPROTEIN IB ALPHA CHAIN, ... | | Authors: | Lechtenberg, B.C, Freund, S.M.V, Huntington, J.A. | | Deposit date: | 2013-11-28 | | Release date: | 2013-12-11 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Gpibalpha Interacts Exclusively with Exosite II of Thrombin
J.Mol.Biol., 426, 2014
|
|
4CH8
 
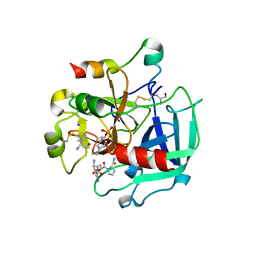 | | High-salt crystal structure of a thrombin-GpIbalpha peptide complex | | Descriptor: | D-phenylalanyl-N-[(2S,3S)-6-{[amino(iminio)methyl]amino}-1-chloro-2-hydroxyhexan-3-yl]-L-prolinamide, GLYCEROL, PLATELET GLYCOPROTEIN IB ALPHA CHAIN, ... | | Authors: | Lechtenberg, B.C, Freund, S.M.V, Huntington, J.A. | | Deposit date: | 2013-11-29 | | Release date: | 2013-12-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Gpibalpha Interacts Exclusively with Exosite II of Thrombin
J.Mol.Biol., 426, 2014
|
|
4LQW
 
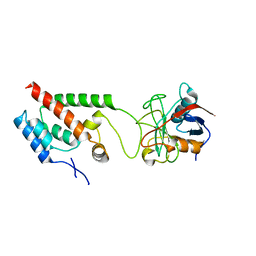 | |
5LJM
 
 | | Structure of SPATA2 PUB domain | | Descriptor: | GLYCEROL, Spermatogenesis-associated protein 2 | | Authors: | Elliott, P.R, Komander, D. | | Deposit date: | 2016-07-18 | | Release date: | 2016-08-24 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.454 Å) | | Cite: | SPATA2 Links CYLD to LUBAC, Activates CYLD, and Controls LUBAC Signaling.
Mol.Cell, 63, 2016
|
|
5M1S
 
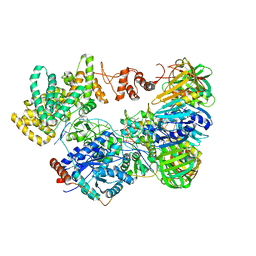 | | Cryo-EM structure of the E. coli replicative DNA polymerase-clamp-exonuclase-theta complex bound to DNA in the editing mode | | Descriptor: | DNA Primer Strand, DNA Template Strand, DNA polymerase III subunit alpha, ... | | Authors: | Fernandez-Leiro, R, Conrad, J, Scheres, S.H.W, Lamers, M.H. | | Deposit date: | 2016-10-10 | | Release date: | 2017-01-18 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (6.7 Å) | | Cite: | Self-correcting mismatches during high-fidelity DNA replication.
Nat. Struct. Mol. Biol., 24, 2017
|
|
6QIK
 
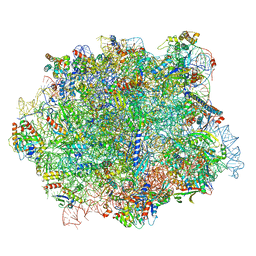 | |
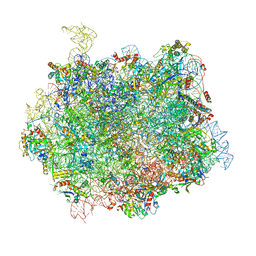
6QT0
 
 | |
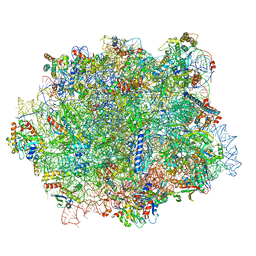
6QTZ
 
 | |
1WIU
 
 | |
1WIT
 
 | |
4OYJ
 
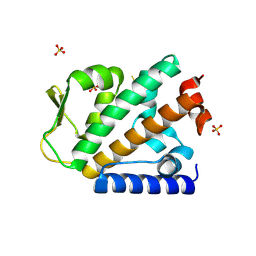 | | Structure of the apo HOIP PUB domain | | Descriptor: | E3 ubiquitin-protein ligase RNF31, SULFATE ION | | Authors: | Elliott, P.R, Komander, D. | | Deposit date: | 2014-02-12 | | Release date: | 2014-05-21 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Molecular Basis and Regulation of OTULIN-LUBAC Interaction.
Mol.Cell, 54, 2014
|
|
4OYK
 
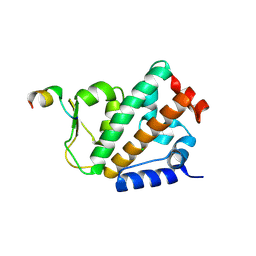 | |
1SRO
 
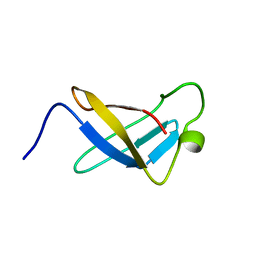 | | S1 RNA BINDING DOMAIN, NMR, 20 STRUCTURES | | Descriptor: | PNPASE | | Authors: | Bycroft, M. | | Deposit date: | 1996-11-27 | | Release date: | 1997-04-01 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The solution structure of the S1 RNA binding domain: a member of an ancient nucleic acid-binding fold.
Cell(Cambridge,Mass.), 88, 1997
|
|
1UTU
 
 | |
4DGE
 
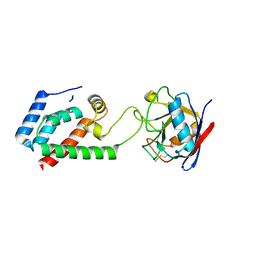 | | TRIMCyp cyclophilin domain from Macaca mulatta: H70C mutant, HIV-1 CA(O-loop) complex | | Descriptor: | TRIMCyp, capsid protein | | Authors: | Caines, M.E.C, Bichel, K, Price, A.J, McEwan, W.A, James, L.C. | | Deposit date: | 2012-01-25 | | Release date: | 2012-02-08 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Diverse HIV viruses are targeted by a conformationally dynamic antiviral.
Nat.Struct.Mol.Biol., 19, 2012
|
|
4DGB
 
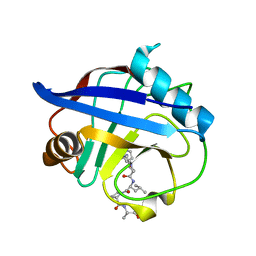 | | TRIMCyp cyclophilin domain from Macaca mulatta: HIV-2 CA cyclophilin-binding loop complex | | Descriptor: | TRIMCyp, capsid protein | | Authors: | Caines, M.E.C, Bichel, K, Price, A.J, McEwan, W.A, James, L.C. | | Deposit date: | 2012-01-25 | | Release date: | 2012-02-08 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.702 Å) | | Cite: | Diverse HIV viruses are targeted by a conformationally dynamic antiviral.
Nat.Struct.Mol.Biol., 19, 2012
|
|
4DGA
 
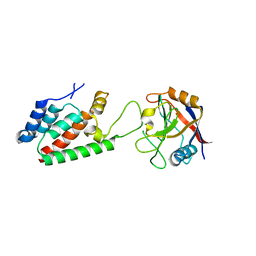 | | TRIMCyp cyclophilin domain from Macaca mulatta: HIV-1 CA(O-loop) complex | | Descriptor: | TRIMCyp, capsid protein | | Authors: | Caines, M.E.C, Bichel, K, Price, A.J, McEwan, W.A, James, L.C. | | Deposit date: | 2012-01-25 | | Release date: | 2012-02-08 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Diverse HIV viruses are targeted by a conformationally dynamic antiviral.
Nat.Struct.Mol.Biol., 19, 2012
|
|
2XWR
 
 | |
2WUK
 
 | |
3SL9
 
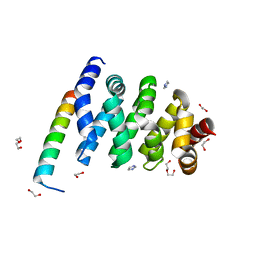 | | X-ray structure of Beta catenin in complex with Bcl9 | | Descriptor: | 1,2-ETHANEDIOL, B-cell CLL/lymphoma 9 protein, Catenin beta-1, ... | | Authors: | Gupta, D, Bienz, M. | | Deposit date: | 2011-06-24 | | Release date: | 2012-02-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | An intrinsically labile alpha-helix abutting the BCL9-binding site of beta-catenin is required for its inhibition by carnosic acid.
Nat Commun, 3, 2012
|
|
3SLA
 
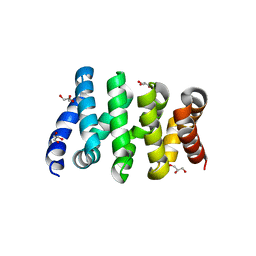 | | X-ray structure of first four repeats of human beta-catenin | | Descriptor: | Catenin beta-1, GLYCEROL, SODIUM ION | | Authors: | Gupta, D, Bienz, M. | | Deposit date: | 2011-06-24 | | Release date: | 2012-02-29 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | An intrinsically labile alpha-helix abutting the BCL9-binding site of beta-catenin is required for its inhibition by carnosic acid.
Nat Commun, 3, 2012
|
|
3VVV
 
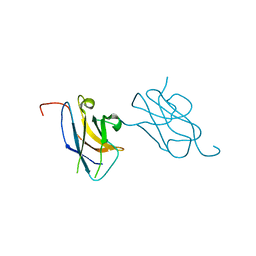 | | Skich domain of NDP52 | | Descriptor: | Calcium-binding and coiled-coil domain-containing protein 2 | | Authors: | Akutsu, M, Muhlinen, N.V, Randow, F, Komander, D. | | Deposit date: | 2012-07-28 | | Release date: | 2013-02-27 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | LC3C, bound selectively by a noncanonical LIR motif in NDP52, is required for antibacterial autophagy
Mol.Cell, 48, 2012
|
|
