6FXM
 
 | | Crystal Structure of full-length Human Lysyl Hydroxylase LH3 - Cocrystal with Mn2+ | | Descriptor: | 2-OXOGLUTARIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Scietti, L, Chiapparino, A, De Giorgi, F, Fumagalli, M, Khoriauli, L, Nergadze, S, Basu, S, Olieric, V, Banushi, B, Giulotto, E, Gissen, P, Forneris, F. | | Deposit date: | 2018-03-09 | | Release date: | 2018-08-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Molecular architecture of the multifunctional collagen lysyl hydroxylase and glycosyltransferase LH3.
Nat Commun, 9, 2018
|
|
6FXT
 
 | | Crystal Structure of full-length Human Lysyl Hydroxylase LH3 - Cocrystal with Fe2+, Mn2+, UDP-Glc | | Descriptor: | 2-OXOGLUTARIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Scietti, L, Chiapparino, A, De Giorgi, F, Fumagalli, M, Khoriauli, L, Nergadze, S, Basu, S, Olieric, V, Banushi, B, Giulotto, E, Gissen, P, Forneris, F. | | Deposit date: | 2018-03-09 | | Release date: | 2018-08-22 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Molecular architecture of the multifunctional collagen lysyl hydroxylase and glycosyltransferase LH3.
Nat Commun, 9, 2018
|
|
6HHE
 
 | | Crystal structure of the medfly Odorant Binding Protein CcapOBP22/CcapOBP69a | | Descriptor: | 2-(2-METHOXYETHOXY)ETHANOL, Odorant binding protein OBP69a, SULFATE ION | | Authors: | Falchetto, M, Ciossani, G, Nenci, S, Mattevi, A, Gasperi, G, Forneris, F. | | Deposit date: | 2018-08-28 | | Release date: | 2018-12-26 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.516 Å) | | Cite: | Structural and biochemical evaluation of Ceratitis capitata odorant-binding protein 22 affinity for odorants involved in intersex communication.
Insect Mol.Biol., 28, 2019
|
|
5FYD
 
 | | Structural and biochemical insights into 7beta-hydroxysteroid dehydrogenase stereoselectivity | | Descriptor: | GLYCEROL, OXIDOREDUCTASE, SHORT CHAIN DEHYDROGENASE/REDUCTASE FAMILY PROTEIN | | Authors: | Savino, S, Ferrandi, E, Forneris, F, Rovida, S, Riva, S, Monti, D, Mattevi, A. | | Deposit date: | 2016-03-07 | | Release date: | 2016-04-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural and Biochemical Insights Into 7Beta-Hydroxysteroid Dehydrogenase Stereoselectivity.
Proteins, 84, 2016
|
|
2WB8
 
 | | Crystal structure of Haspin kinase | | Descriptor: | SERINE/THREONINE-PROTEIN KINASE HASPIN | | Authors: | Villa, F, Tortorci, M, Forneris, F, Mattevi, A, Musacchio, A. | | Deposit date: | 2009-02-23 | | Release date: | 2009-11-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal Structure of the Catalytic Domain of Haspin, an Atypical Kinase Implicated in Chromatin Organization.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
4A5W
 
 | | Crystal structure of C5b6 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, COMPLEMENT C5, ... | | Authors: | Hadders, M.A, Bubeck, D, Forneris, F, Pangburn, M, Llorca, O, Lea, S.M, Gros, P. | | Deposit date: | 2011-10-28 | | Release date: | 2012-03-14 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Assembly and Regulation of the Membrane Attack Complex Based on Structures of C5B6 and Sc5B9.
Cell Rep., 1, 2012
|
|
4CDJ
 
 | | Structure of ZNRF3 ectodomain | | Descriptor: | 1,2-ETHANEDIOL, E3 UBIQUITIN-PROTEIN LIGASE ZNRF3, FORMIC ACID | | Authors: | Peng, W.C, de Lau, W, Madoori, P.K, Forneris, F, Granneman, J.C.M, Clevers, H, Gros, P. | | Deposit date: | 2013-11-01 | | Release date: | 2014-01-08 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structures of Wnt-Antagonist Znrf3 and its Complex with R-Spondin 1 and Implications for Signaling.
Plos One, 8, 2013
|
|
4CDK
 
 | | Structure of ZNRF3-RSPO1 | | Descriptor: | E3 UBIQUITIN-PROTEIN LIGASE ZNRF3, R-SPONDIN-1 | | Authors: | Peng, W.C, de Lau, W, Madoori, P.K, Forneris, F, Granneman, J.C.M, Clevers, H, Gros, P. | | Deposit date: | 2013-11-01 | | Release date: | 2014-01-08 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structures of Wnt-Antagonist Znrf3 and its Complex with R-Spondin 1 and Implications for Signaling.
Plos One, 8, 2013
|
|
4BSP
 
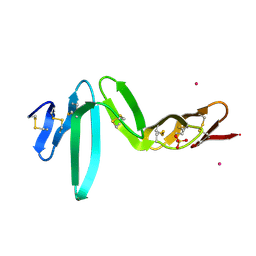 | | Crystal structure of R-spondin 1 (Fu1Fu2) - Holmium soak | | Descriptor: | HOLMIUM ATOM, R-SPONDIN-1, SULFATE ION | | Authors: | Peng, W.C, de Lau, W, Forneris, F, Granneman, J.C.M, Huch, M, Clevers, H, Gros, P. | | Deposit date: | 2013-06-11 | | Release date: | 2013-06-19 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of Stem Cell Growth Factor R-Spondin 1 in Complex with the Ectodomain of its Receptor Lgr5.
Cell Rep., 3, 2013
|
|
4BSO
 
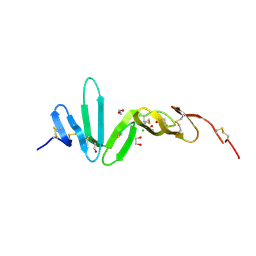 | | Crystal structure of R-spondin 1 (Fu1Fu2) - Native | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, R-SPONDIN-1 | | Authors: | Peng, W.C, de Lau, W, Forneris, F, Granneman, J.C.M, Huch, M, Clevers, H, Gros, P. | | Deposit date: | 2013-06-11 | | Release date: | 2013-06-19 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of Stem Cell Growth Factor R-Spondin 1 in Complex with the Ectodomain of its Receptor Lgr5.
Cell Rep., 3, 2013
|
|
4BST
 
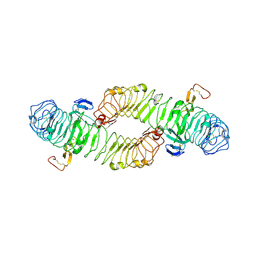 | | Structure of the ectodomain of LGR5 in complex with R-spondin-1 (Fu1Fu2) in P6122 crystal form | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, LEUCINE-RICH REPEAT-CONTAINING G-PROTEIN COUPLED RECEPTOR 5, R-SPONDIN-1, ... | | Authors: | Peng, W.C, de Lau, W, Forneris, F, Granneman, J.C.M, Huch, M, Clevers, H, Gros, P. | | Deposit date: | 2013-06-11 | | Release date: | 2013-06-19 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (4.3 Å) | | Cite: | Structure of Stem Cell Growth Factor R-Spondin 1 in Complex with the Ectodomain of its Receptor Lgr5.
Cell Rep., 3, 2013
|
|
4BSS
 
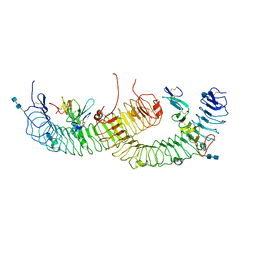 | | Structure of the ectodomain of LGR5 in complex with R-spondin-1 (Fu1Fu2) in P21 crystal form | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, LEUCINE-RICH REPEAT-CONTAINING G-PROTEIN COUPLED RECEPTOR 5, ... | | Authors: | Peng, W.C, de Lau, W, Forneris, F, Granneman, J.C.M, Huch, M, Clevers, H, Gros, P. | | Deposit date: | 2013-06-11 | | Release date: | 2013-06-19 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structure of Stem Cell Growth Factor R-Spondin 1 in Complex with the Ectodomain of its Receptor Lgr5.
Cell Rep., 3, 2013
|
|
4BSU
 
 | | Structure of the ectodomain of LGR5 in complex with R-spondin-1 (Fu1Fu2) in C2 crystal form | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, LEUCINE-RICH REPEAT-CONTAINING G-PROTEIN COUPLED RECEPTOR 5, ... | | Authors: | Peng, W.C, de Lau, W, Forneris, F, Granneman, J.C.M, Huch, M, Clevers, H, Gros, P. | | Deposit date: | 2013-06-11 | | Release date: | 2013-06-26 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structure of Stem Cell Growth Factor R-Spondin 1 in Complex with the Ectodomain of its Receptor Lgr5.
Cell Rep., 3, 2013
|
|
4BSR
 
 | | Structure of the ectodomain of LGR5 in complex with R-spondin-1 (Fu1Fu2) in P22121 crystal form | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, LEUCINE-RICH REPEAT-CONTAINING G-PROTEIN COUPLED RECEPTOR 5, ... | | Authors: | Peng, W.C, de Lau, W, Forneris, F, Granneman, J.C.M, Huch, M, Clevers, H, Gros, P. | | Deposit date: | 2013-06-11 | | Release date: | 2013-06-19 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structure of Stem Cell Growth Factor R-Spondin 1 in Complex with the Ectodomain of its Receptor Lgr5.
Cell Rep., 3, 2013
|
|
2X0L
 
 | | Crystal structure of a neuro-specific splicing variant of human histone lysine demethylase LSD1. | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, HISTONE H3 PEPTIDE, LYSINE-SPECIFIC HISTONE DEMETHYLASE 1, ... | | Authors: | Zibetti, C, Adamo, A, Binda, C, Forneris, F, Verpelli, C, Ginelli, E, Mattevi, A, Sala, C, Battaglioli, E. | | Deposit date: | 2009-12-15 | | Release date: | 2010-03-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Alternative Splicing of the Histone Demethylase Lsd1/Kdm1 Contributes to the Modulation of Neurite Morphogenesis in the Mammalian Nervous System.
J.Neurosci., 30, 2010
|
|
2XAJ
 
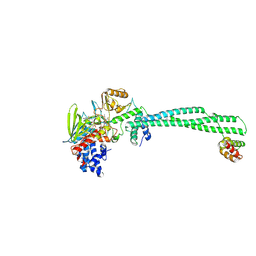 | | Crystal structure of LSD1-CoREST in complex with (-)-trans-2- phenylcyclopropyl-1-amine | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, LYSINE-SPECIFIC HISTONE DEMETHYLASE 1, PHENYLETHYLENECARBOXYLIC ACID, ... | | Authors: | Binda, C, Valente, S, Romanenghi, M, Pilotto, S, Cirilli, R, Karytinos, A, Ciossani, G, Botrugno, O.A, Forneris, F, Tardugno, M, Edmondson, D.E, Minucci, S, Mattevi, A, Mai, A. | | Deposit date: | 2010-03-31 | | Release date: | 2010-05-05 | | Last modified: | 2025-03-12 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Biochemical, Structural, and Biological Evaluation of Tranylcypromine Derivatives as Inhibitors of Histone Demethylases Lsd1 and Lsd2.
J.Am.Chem.Soc., 132, 2010
|
|
2XAF
 
 | | Crystal structure of LSD1-CoREST in complex with para-bromo-(+)-cis-2- phenylcyclopropyl-1-amine | | Descriptor: | 3-(4-BROMOPHENYL)PROPANAMIDE, FLAVIN-ADENINE DINUCLEOTIDE, LYSINE-SPECIFIC HISTONE DEMETHYLASE 1, ... | | Authors: | Binda, C, Valente, S, Romanenghi, M, Pilotto, S, Cirilli, R, Karytinos, A, Ciossani, G, Botrugno, O.A, Forneris, F, Tardugno, M, Edmondson, D.E, Minucci, S, Mattevi, A, Mai, A. | | Deposit date: | 2010-03-31 | | Release date: | 2010-05-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Biochemical, Structural, and Biological Evaluation of Tranylcypromine Derivatives as Inhibitors of Histone Demethylases Lsd1 and Lsd2.
J.Am.Chem.Soc., 132, 2010
|
|
2XAQ
 
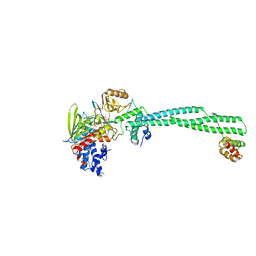 | | Crystal structure of LSD1-CoREST in complex with a tranylcypromine derivative (MC2584, 13b) | | Descriptor: | 3-{4-[(PHENYLCARBONYL)AMINO]PHENYL}PROPANOIC ACID, FLAVIN-ADENINE DINUCLEOTIDE, LYSINE-SPECIFIC HISTONE DEMETHYLASE 1, ... | | Authors: | Binda, C, Valente, S, Romanenghi, M, Pilotto, S, Cirilli, R, Karytinos, A, Ciossani, G, Botrugno, O.A, Forneris, F, Tardugno, M, Edmondson, D.E, Minucci, S, Mattevi, A, Mai, A. | | Deposit date: | 2010-03-31 | | Release date: | 2010-05-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Biochemical, Structural, and Biological Evaluation of Tranylcypromine Derivatives as Inhibitors of Histone Demethylases Lsd1 and Lsd2.
J.Am.Chem.Soc., 132, 2010
|
|
2XAH
 
 | | Crystal structure of LSD1-CoREST in complex with (+)-trans-2- phenylcyclopropyl-1-amine | | Descriptor: | 3-PHENYLPROPANAL, FLAVIN-ADENINE DINUCLEOTIDE, LYSINE-SPECIFIC HISTONE DEMETHYLASE 1, ... | | Authors: | Binda, C, Valente, S, Romanenghi, M, Pilotto, S, Cirilli, R, Karytinos, A, Ciossani, G, Botrugno, O.A, Forneris, F, Tardugno, M, Edmondson, D.E, Minucci, S, Mattevi, A, Mai, A. | | Deposit date: | 2010-03-31 | | Release date: | 2010-05-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Biochemical, Structural, and Biological Evaluation of Tranylcypromine Derivatives as Inhibitors of Histone Demethylases Lsd1 and Lsd2.
J.Am.Chem.Soc., 132, 2010
|
|
2XAS
 
 | | Crystal structure of LSD1-CoREST in complex with a tranylcypromine derivative (MC2580, 14e) | | Descriptor: | 3-[4-({N-[(BENZYLOXY)CARBONYL]-L-PHENYLALANYL}AMINO)PHENYL]PROPANOIC ACID, FLAVIN-ADENINE DINUCLEOTIDE, LYSINE-SPECIFIC HISTONE DEMETHYLASE 1, ... | | Authors: | Binda, C, Valente, S, Romanenghi, M, Pilotto, S, Cirilli, R, Karytinos, A, Ciossani, G, Botrugno, O.A, Forneris, F, Tardugno, M, Edmondson, D.E, Minucci, S, Mattevi, A, Mai, A. | | Deposit date: | 2010-03-31 | | Release date: | 2010-05-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Biochemical, Structural, and Biological Evaluation of Tranylcypromine Derivatives as Inhibitors of Histone Demethylases Lsd1 and Lsd2.
J.Am.Chem.Soc., 132, 2010
|
|
2XAG
 
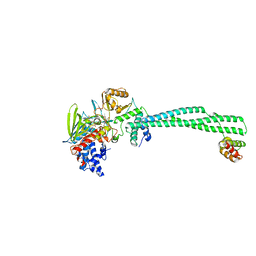 | | Crystal structure of LSD1-CoREST in complex with para-bromo-(-)-trans- 2-phenylcyclopropyl-1-amine | | Descriptor: | 3-(4-BROMOPHENYL)PROPANAMIDE, FLAVIN-ADENINE DINUCLEOTIDE, LYSINE-SPECIFIC HISTONE DEMETHYLASE 1, ... | | Authors: | Binda, C, Valente, S, Romanenghi, M, Pilotto, S, Cirilli, R, Karytinos, A, Ciossani, G, Botrugno, O.A, Forneris, F, Tardugno, M, Edmondson, D.E, Minucci, S, Mattevi, A, Mai, A. | | Deposit date: | 2010-03-31 | | Release date: | 2010-05-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Biochemical, Structural, and Biological Evaluation of Tranylcypromine Derivatives as Inhibitors of Histone Demethylases Lsd1 and Lsd2.
J.Am.Chem.Soc., 132, 2010
|
|
8D1B
 
 | |
7OCM
 
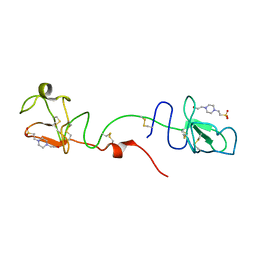 | | K1K1H6, a potent recombinant minimal hepatocyte growth factor/scatter factor mimic | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Hepatocyte growth factor alpha chain,Hepatocyte growth factor alpha chain | | Authors: | de Jonge, H, de Nola, G, Gherardi, E. | | Deposit date: | 2021-04-27 | | Release date: | 2021-07-07 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Dimerization of kringle 1 domain from hepatocyte growth factor/scatter factor provides a potent MET receptor agonist.
Life Sci Alliance, 5, 2022
|
|
7OCL
 
 | |
1OVU
 
 | |
