6PPO
 
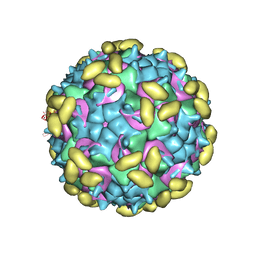 | | Rhinovirus C15 complexed with domain I of receptor CDHR3 | | Descriptor: | CALCIUM ION, Cadherin-related family member 3, Capsid protein VP1, ... | | Authors: | Sun, Y, Watters, K, Klose, T, Palmenberg, A.C. | | Deposit date: | 2019-07-08 | | Release date: | 2020-03-11 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM structure of rhinovirus C15a bound to its cadherin-related protein 3 receptor.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
8IV5
 
 | | Cryo-EM structure of SARS-CoV-2 spike protein in complex with double nAbs 8H12 and 1C4 (local refinement) | | Descriptor: | Spike protein S1, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, heavy chain of 1C4, ... | | Authors: | Sun, H, Jiang, Y, Zheng, Q, Li, S, Xia, N. | | Deposit date: | 2023-03-26 | | Release date: | 2023-08-16 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (3.77 Å) | | Cite: | Two antibodies show broad, synergistic neutralization against SARS-CoV-2 variants by inducing conformational change within the RBD.
Protein Cell, 15, 2024
|
|
8IV8
 
 | | Cryo-EM structure of SARS-CoV-2 spike protein in complex with double nAbs 3E2 and 1C4 (local refinement) | | Descriptor: | Spike protein S1, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, heavy chain of 1C4, ... | | Authors: | Sun, H, Jiang, Y, Zheng, Q, Li, S, Xia, N. | | Deposit date: | 2023-03-26 | | Release date: | 2023-08-16 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (3.92 Å) | | Cite: | Two antibodies show broad, synergistic neutralization against SARS-CoV-2 variants by inducing conformational change within the RBD.
Protein Cell, 15, 2024
|
|
8IVA
 
 | | Cryo-EM structure of SARS-CoV-2 spike protein in complex with double nAbs XMA01 and 3E2 (local refinement) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1, heavy chain of 3E2, ... | | Authors: | Sun, H, Jiang, Y, Zheng, Q, Li, S, Xia, N. | | Deposit date: | 2023-03-26 | | Release date: | 2023-08-16 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (3.95 Å) | | Cite: | Two antibodies show broad, synergistic neutralization against SARS-CoV-2 variants by inducing conformational change within the RBD.
Protein Cell, 15, 2024
|
|
8IV4
 
 | | Cryo-EM structure of SARS-CoV-2 spike protein in complex with double nAbs 8H12 and 3E2 (local refinement) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1, heavy chain of 3E2, ... | | Authors: | Sun, H, Jiang, Y, Zheng, Q, Li, S, Xia, N. | | Deposit date: | 2023-03-26 | | Release date: | 2023-08-16 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (3.59 Å) | | Cite: | Two antibodies show broad, synergistic neutralization against SARS-CoV-2 variants by inducing conformational change within the RBD.
Protein Cell, 15, 2024
|
|
6PSF
 
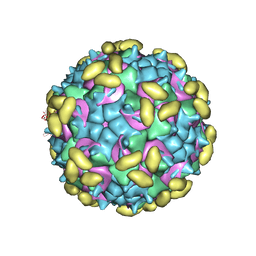 | | Rhinovirus C15 complexed with domains I and II of receptor CDHR3 | | Descriptor: | Cadherin-related family member 3, Capsid protein VP1, Capsid protein VP2, ... | | Authors: | Sun, Y, Watters, K, Klose, T, Palmenberg, A.C. | | Deposit date: | 2019-07-12 | | Release date: | 2020-03-11 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Cryo-EM structure of rhinovirus C15a bound to its cadherin-related protein 3 receptor.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
7KTR
 
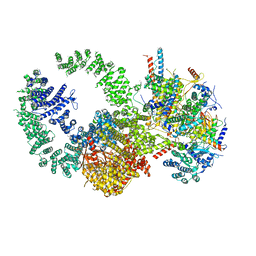 | | Cryo-EM structure of the human SAGA coactivator complex (TRRAP, core) | | Descriptor: | Ataxin-7, INOSITOL HEXAKISPHOSPHATE, Isoform 3 of Transcription factor SPT20 homolog, ... | | Authors: | Herbst, D.A, Esbin, M.N, Nogales, E. | | Deposit date: | 2020-11-24 | | Release date: | 2021-11-10 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.93 Å) | | Cite: | Structure of the human SAGA coactivator complex.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7KTS
 
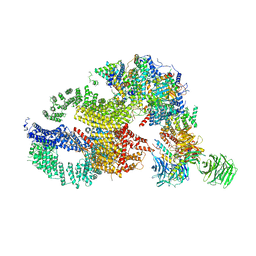 | | Negative stain EM structure of the human SAGA coactivator complex (TRRAP, core, splicing module) | | Descriptor: | Ataxin-7, Isoform 3 of Transcription factor SPT20 homolog, STAGA complex 65 subunit gamma, ... | | Authors: | Herbst, D.A, Esbin, M.N, Nogales, E. | | Deposit date: | 2020-11-24 | | Release date: | 2021-11-10 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (19.09 Å) | | Cite: | Structure of the human SAGA coactivator complex.
Nat.Struct.Mol.Biol., 28, 2021
|
|
8WGV
 
 | | BA.2(S375) Spike (S6P)/hACE2 complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Wei, X, Zhang, Z. | | Deposit date: | 2023-09-22 | | Release date: | 2024-02-28 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.92 Å) | | Cite: | The receptor binding domain of SARS-CoV-2 Omicron subvariants targets Siglec-9 to decrease its immunogenicity by preventing macrophage phagocytosis.
Nat.Immunol., 25, 2024
|
|
8WGW
 
 | | Local refinement of RBD-ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Wei, X, Zhang, Z. | | Deposit date: | 2023-09-22 | | Release date: | 2024-02-28 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | The receptor binding domain of SARS-CoV-2 Omicron subvariants targets Siglec-9 to decrease its immunogenicity by preventing macrophage phagocytosis.
Nat.Immunol., 25, 2024
|
|
8T9R
 
 | |
8T1X
 
 | |
4PSX
 
 | | Crystal structure of histone acetyltransferase complex | | Descriptor: | COENZYME A, Histone H3, Histone H4, ... | | Authors: | Yang, M, Li, Y. | | Deposit date: | 2014-03-08 | | Release date: | 2014-07-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.509 Å) | | Cite: | Hat2p recognizes the histone H3 tail to specify the acetylation of the newly synthesized H3/H4 heterodimer by the Hat1p/Hat2p complex
Genes Dev., 28, 2014
|
|
4PH4
 
 | | The crystal structure of Human VPS34 in complex with PIK-III | | Descriptor: | 4'-(cyclopropylmethyl)-N~2~-(pyridin-4-yl)-4,5'-bipyrimidine-2,2'-diamine, GLYCEROL, Phosphatidylinositol 3-kinase catalytic subunit type 3 | | Authors: | Knapp, M.S, Elling, R.A. | | Deposit date: | 2014-05-04 | | Release date: | 2014-10-29 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Selective VPS34 inhibitor blocks autophagy and uncovers a role for NCOA4 in ferritin degradation and iron homeostasis in vivo.
Nat.Cell Biol., 16, 2014
|
|
4PSW
 
 | | Crystal structure of histone acetyltransferase complex | | Descriptor: | COENZYME A, Histone H4 type VIII, Histone acetyltransferase type B catalytic subunit, ... | | Authors: | Yang, M, Li, Y. | | Deposit date: | 2014-03-08 | | Release date: | 2014-07-09 | | Method: | X-RAY DIFFRACTION (2.101 Å) | | Cite: | Hat2p recognizes the histone H3 tail to specify the acetylation of the newly synthesized H3/H4 heterodimer by the Hat1p/Hat2p complex
Genes Dev., 28, 2014
|
|
7YJ1
 
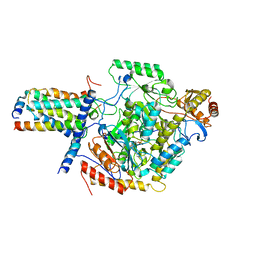 | | Cryo-EM structure of SPT-ORMDL3 (ORMDL3-deltaN2) complex | | Descriptor: | ORM1-like protein 3, PYRIDOXAL-5'-PHOSPHATE, Serine palmitoyltransferase 1, ... | | Authors: | Xie, T, Liu, P, Gong, X. | | Deposit date: | 2022-07-19 | | Release date: | 2023-07-05 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Ceramide sensing by human SPT-ORMDL complex for establishing sphingolipid homeostasis.
Nat Commun, 14, 2023
|
|
7YIU
 
 | | Cryo-EM structure of the C6-ceramide-bound SPT-ORMDL3 complex | | Descriptor: | HEXADECANE, N-((E,2S,3R)-1,3-DIHYDROXYOCTADEC-4-EN-2-YL)HEXANAMIDE, ORM1-like protein 3, ... | | Authors: | Xie, T, Liu, P, Gong, X. | | Deposit date: | 2022-07-18 | | Release date: | 2023-07-05 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Ceramide sensing by human SPT-ORMDL complex for establishing sphingolipid homeostasis.
Nat Commun, 14, 2023
|
|
7YJ2
 
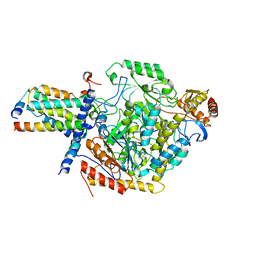 | | Cryo-EM structure of SPT-ORMDL3 (ORMDL3-N13A) complex | | Descriptor: | ORM1-like protein 3, PYRIDOXAL-5'-PHOSPHATE, Serine palmitoyltransferase 1, ... | | Authors: | Xie, T, Liu, P, Gong, X. | | Deposit date: | 2022-07-19 | | Release date: | 2023-07-05 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Ceramide sensing by human SPT-ORMDL complex for establishing sphingolipid homeostasis.
Nat Commun, 14, 2023
|
|
7YIY
 
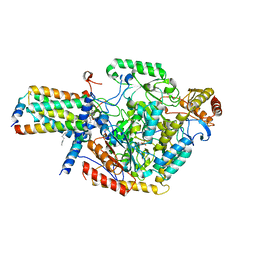 | | Cryo-EM structure of SPT-ORMDL3 complex | | Descriptor: | N-[(2S,3R,4E)-1,3-dihydroxyoctadec-4-en-2-yl]tetracosanamide, ORM1-like protein 3, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Xie, T, Liu, P, Gong, X. | | Deposit date: | 2022-07-18 | | Release date: | 2023-07-05 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Ceramide sensing by human SPT-ORMDL complex for establishing sphingolipid homeostasis.
Nat Commun, 14, 2023
|
|
7E7Q
 
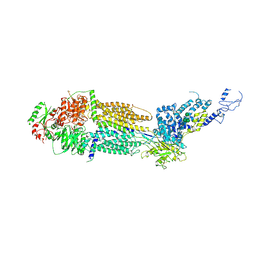 | | Cryo-EM structure of human ABCA4 in ATP-bound state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Xie, T, Zhang, Z.K, Gong, X. | | Deposit date: | 2021-02-26 | | Release date: | 2021-06-30 | | Last modified: | 2021-07-07 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis of substrate recognition and translocation by human ABCA4.
Nat Commun, 12, 2021
|
|
7E7I
 
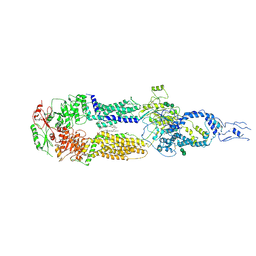 | | Cryo-EM structure of human ABCA4 in the apo state | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, 2-acetamido-2-deoxy-beta-D-glucopyranose, Retinal-specific phospholipid-transporting ATPase ABCA4, ... | | Authors: | Xie, T, Zhang, Z.K, Gong, X. | | Deposit date: | 2021-02-26 | | Release date: | 2021-06-30 | | Last modified: | 2021-07-07 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis of substrate recognition and translocation by human ABCA4.
Nat Commun, 12, 2021
|
|
7E7O
 
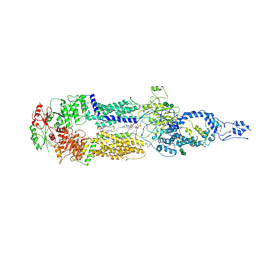 | | Cryo-EM structure of human ABCA4 in NRPE-bound state | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, 2-acetamido-2-deoxy-beta-D-glucopyranose, Retinal-specific phospholipid-transporting ATPase ABCA4, ... | | Authors: | Xie, T, Zhang, Z.K, Gong, X. | | Deposit date: | 2021-02-26 | | Release date: | 2021-06-30 | | Last modified: | 2021-07-07 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural basis of substrate recognition and translocation by human ABCA4.
Nat Commun, 12, 2021
|
|
7W02
 
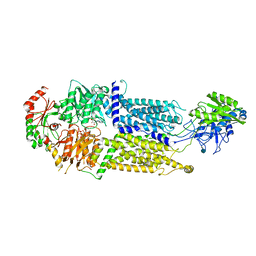 | | Cryo-EM structure of ATP-bound ABCA3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Xie, T, Zhang, Z.K, Yue, J, Gong, X. | | Deposit date: | 2021-11-17 | | Release date: | 2022-04-20 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM structures of the human surfactant lipid transporter ABCA3.
Sci Adv, 8, 2022
|
|
7W01
 
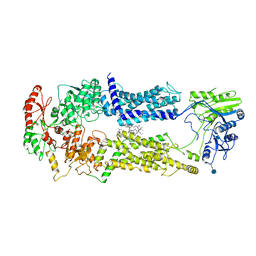 | | Cryo-EM structure of nucleotide-free ABCA3 | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 1,2-DIMYRISTOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Xie, T, Zhang, Z.K, Yue, J, Gong, X. | | Deposit date: | 2021-11-17 | | Release date: | 2022-04-27 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM structures of the human surfactant lipid transporter ABCA3.
Sci Adv, 8, 2022
|
|
7F6L
 
 | | Crystal structure of human MUS81-EME2 complex | | Descriptor: | Crossover junction endonuclease MUS81, Probable crossover junction endonuclease EME2 | | Authors: | Hua, Z.K, Zhang, D.P, Yuan, C, Lin, Z.H. | | Deposit date: | 2021-06-25 | | Release date: | 2022-03-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structure of the human MUS81-EME2 complex.
Structure, 30, 2022
|
|
