6LDH
 
 | |
4GPD
 
 | |
4CHI
 
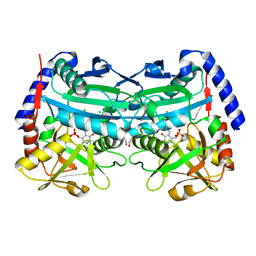 | | (R)-selective amine transaminase from Aspergillus fumigatus at 1.27 A resolution | | Descriptor: | BRANCHED-CHAIN AMINO ACID AMINOTRANSFERASE, CHLORIDE ION, GLYCEROL, ... | | Authors: | Thomsen, M, Palm, G.J, Hinrichs, W. | | Deposit date: | 2013-12-02 | | Release date: | 2014-04-30 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (1.27 Å) | | Cite: | Crystallographic Characterization of the (R)-Selective Amine Transaminase from Aspergillus Fumigatus.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
8LDH
 
 | |
4UUG
 
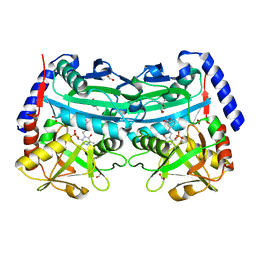 | | The (R)-selective amine transaminase from Aspergillus fumigatus with inhibitor bound | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, 3-[O-PHOSPHONOPYRIDOXYL]--AMINO-BENZOIC ACID, ... | | Authors: | Thomsen, M, Hinrichs, W. | | Deposit date: | 2014-07-28 | | Release date: | 2014-11-26 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural and Biochemical Characterization of the Dual Substrate Recognition of the (R)-Selective Amine Transaminase from Aspergillus Fumigatus
FEBS J., 282, 2015
|
|
1LDM
 
 | |
8ROY
 
 | | Structure of the human DDB1-DDA1-DCAF15 E3 ubiquitin ligase bound to compound furan 24 | | Descriptor: | 1-[5-[[3,4-bis(chloranyl)-1~{H}-indol-7-yl]sulfamoyl]-3-methyl-furan-2-yl]carbonyl-~{N}-methyl-piperidine-4-carboxamide, DDB1- and CUL4-associated factor 15, DET1- and DDB1-associated protein 1, ... | | Authors: | Shilliday, F, Lucas, S.C.C, Richter, M, Michaelides, I.N, Fusani, L. | | Deposit date: | 2024-01-12 | | Release date: | 2024-04-03 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Optimization of Potent Ligands for the E3 Ligase DCAF15 and Evaluation of Their Use in Heterobifunctional Degraders.
J.Med.Chem., 67, 2024
|
|
8ROX
 
 | | Structure of the human DDB1-DDA1-DCAF15 E3 ubiquitin ligase bound to compound furan 12 | | Descriptor: | 5-[[3,4-bis(chloranyl)-1~{H}-indol-7-yl]sulfamoyl]-~{N},~{N},3-trimethyl-furan-2-carboxamide;ethane, DDB1- and CUL4-associated factor 15, DET1- and DDB1-associated protein 1, ... | | Authors: | Shilliday, F, Lucas, S.C.C, Richter, M, Michaelides, I.N, Fusani, L. | | Deposit date: | 2024-01-12 | | Release date: | 2024-04-03 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Optimization of Potent Ligands for the E3 Ligase DCAF15 and Evaluation of Their Use in Heterobifunctional Degraders.
J.Med.Chem., 67, 2024
|
|
4HT1
 
 | | Human TWEAK in complex with the Fab fragment of a neutralizing antibody | | Descriptor: | Tumor necrosis factor ligand superfamily member 12, chimeric antibody Fab | | Authors: | Lammens, A. | | Deposit date: | 2012-10-31 | | Release date: | 2013-06-12 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.498 Å) | | Cite: | Crystal Structure of Human TWEAK in Complex with the Fab Fragment of a Neutralizing Antibody Reveals Insights into Receptor Binding.
Plos One, 8, 2013
|
|
5KIC
 
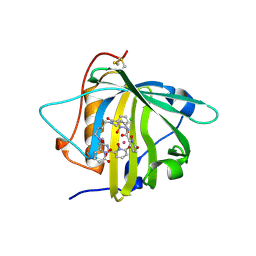 | | Long-sought stabilization of berkelium(IV) in solution: An anomaly within the heavy actinide series | | Descriptor: | CALIFORNIUM ION, CHLORIDE ION, N,N'-butane-1,4-diylbis[1-hydroxy-N-(3-{[(1-hydroxy-6-oxo-1,6-dihydropyridin-2-yl)carbonyl]amino}propyl)-6-oxo-1,6-dihydropyridine-2-carboxamide], ... | | Authors: | Rupert, P.B, Strong, R.K. | | Deposit date: | 2016-06-16 | | Release date: | 2017-04-26 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Chelation and stabilization of berkelium in oxidation state +IV.
Nat Chem, 9, 2017
|
|
6XK9
 
 | | Cereblon in complex with DDB1, CC-90009, and GSPT1 | | Descriptor: | 2-(4-chlorophenyl)-N-({2-[(3S)-2,6-dioxopiperidin-3-yl]-1-oxo-2,3-dihydro-1H-isoindol-5-yl}methyl)-2,2-difluoroacetamide, DNA damage-binding protein 1, Eukaryotic peptide chain release factor GTP-binding subunit ERF3A, ... | | Authors: | Clayton, T.L, Tran, E.T, Zhu, J, Pagarigan, B.E, Matyskiela, M.E, Chamberlain, P.P. | | Deposit date: | 2020-06-25 | | Release date: | 2020-12-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.64 Å) | | Cite: | CC-90009, a novel cereblon E3 ligase modulator, targets acute myeloid leukemia blasts and leukemia stem cells.
Blood, 137, 2021
|
|
7DE8
 
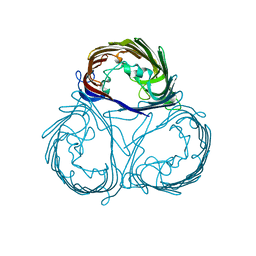 | |
3G3T
 
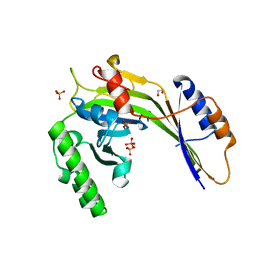 | | Crystal structure of a eukaryotic polyphosphate polymerase in complex with orthophosphate | | Descriptor: | 1,2-ETHANEDIOL, PHOSPHATE ION, Vacuolar transporter chaperone 4 | | Authors: | Lenherr, E.D, Hothorn, M, Scheffzek, K. | | Deposit date: | 2009-02-02 | | Release date: | 2009-05-05 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Catalytic core of a membrane-associated eukaryotic polyphosphate polymerase.
Science, 324, 2009
|
|
6ZK6
 
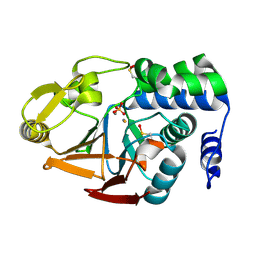 | | Protein Phosphatase 1 (PP1) T320E mutant | | Descriptor: | FE (III) ION, MANGANESE (II) ION, PHOSPHATE ION, ... | | Authors: | Salvi, F, Barabas, O, Koehn, M. | | Deposit date: | 2020-06-29 | | Release date: | 2020-11-18 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Towards Dissecting the Mechanism of Protein Phosphatase-1 Inhibition by Its C-Terminal Phosphorylation.
Chembiochem, 22, 2021
|
|
3G3R
 
 | |
3G3Q
 
 | |
4G9J
 
 | | Protein Ser/Thr phosphatase-1 in complex with cell-permeable peptide | | Descriptor: | MANGANESE (II) ION, Serine/threonine-protein phosphatase PP1-alpha catalytic subunit, synthetic peptide | | Authors: | Sukackaite, R, Chatterjee, J, Beullens, M, Bollen, M, Koehn, M, Hart, D.J. | | Deposit date: | 2012-07-24 | | Release date: | 2012-09-19 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Development of a Peptide that Selectively Activates Protein Phosphatase-1 in Living Cells.
Angew.Chem.Int.Ed.Engl., 51, 2012
|
|
7B0B
 
 | | Fab HbnC3t1p1_C6 bound to SARS-CoV-2 RBD | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Fab heavy chain, IGK@ protein, ... | | Authors: | Borenstein-Katz, A, Diskin, R. | | Deposit date: | 2020-11-19 | | Release date: | 2021-12-01 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.98 Å) | | Cite: | Somatic hypermutation introduces bystander mutations that prepare SARS-CoV-2 antibodies for emerging variants.
Immunity, 56, 2023
|
|
3G3O
 
 | |
3G3U
 
 | |
6S8I
 
 | | Structure of ZEBOV GP in complex with 3T0265 antibody | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein, Enveloped Glycoprotein 1, ... | | Authors: | Diskin, R, Cohen-Dvashi, H. | | Deposit date: | 2019-07-10 | | Release date: | 2020-02-12 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.99 Å) | | Cite: | Structural Basis for a Convergent Immune Response against Ebola Virus.
Cell Host Microbe, 27, 2020
|
|
6S8D
 
 | | Structure of ZEBOV GP in complex with 1T0227 antibody | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein, Heavy chain, ... | | Authors: | Diskin, R, Cohen-Dvashi, H. | | Deposit date: | 2019-07-09 | | Release date: | 2020-02-12 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.49 Å) | | Cite: | Structural Basis for a Convergent Immune Response against Ebola Virus.
Cell Host Microbe, 27, 2020
|
|
6GMH
 
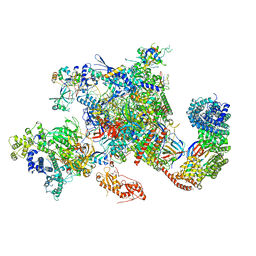 | | Structure of activated transcription complex Pol II-DSIF-PAF-SPT6 | | Descriptor: | CDC73, CTR9,RNA polymerase-associated protein CTR9 homolog,RNA polymerase-associated protein CTR9 homolog, DNA-directed RNA polymerase II subunit RPB9, ... | | Authors: | Vos, S.M, Farnung, L, Boehing, M, Linden, A, Wigge, C, Urlaub, H, Cramer, P. | | Deposit date: | 2018-05-26 | | Release date: | 2018-08-22 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structure of activated transcription complex Pol II-DSIF-PAF-SPT6.
Nature, 560, 2018
|
|
6S8J
 
 | | Structure of ZEBOV GP in complex with 5T0180 antibody | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope Glycoprotein 1, Envelope glycoprotein, ... | | Authors: | Diskin, R, Cohen-Dvashi, H. | | Deposit date: | 2019-07-10 | | Release date: | 2020-02-12 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.91 Å) | | Cite: | Structural Basis for a Convergent Immune Response against Ebola Virus.
Cell Host Microbe, 27, 2020
|
|
5AES
 
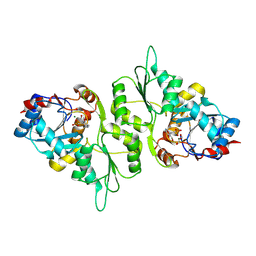 | | Crystal Structure of murine Chronophin (Pyridoxal Phosphate Phosphatase) in Complex with a PNP-derived Inhibitor | | Descriptor: | GLYCEROL, MAGNESIUM ION, PYRIDOXAL PHOSPHATE PHOSPHATASE, ... | | Authors: | Knobloch, G, Jabari, N, Koehn, M, Gohla, A, Schindelin, H. | | Deposit date: | 2015-01-09 | | Release date: | 2015-04-01 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.751 Å) | | Cite: | Synthesis of Hydrolysis-Resistant Pyridoxal 5'-Phosphate Analogs and Their Biochemical and X-Ray Crystallographic Characterization with the Pyridoxal Phosphatase Chronophin.
Bioorg.Med.Chem., 23, 2015
|
|
