5ENR
 
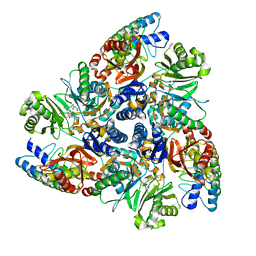 | | MBX3135 bound structure of bacterial efflux pump. | | Descriptor: | DARPin, Multidrug efflux pump subunit AcrB,Multidrug efflux pump subunit AcrB, ~{N}-[4-[2-[[5-cyano-8-[(2~{S},6~{S})-2,6-dimethylmorpholin-4-yl]-3,3-dimethyl-1,4-dihydropyrano[3,4-c]pyridin-6-yl]sulfanyl]ethyl]phenyl]prop-2-enamide | | Authors: | Sjuts, H, Ornik, A.R, Pos, K.M. | | Deposit date: | 2015-11-09 | | Release date: | 2016-04-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Molecular basis for inhibition of AcrB multidrug efflux pump by novel and powerful pyranopyridine derivatives.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5ENO
 
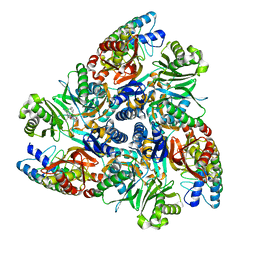 | | MBX2319 bound structure of bacterial efflux pump. | | Descriptor: | 3,3-dimethyl-8-morpholin-4-yl-6-(2-phenylethylsulfanyl)-1,4-dihydropyrano[3,4-c]pyridine-5-carbonitrile, DARPin, Multidrug efflux pump subunit AcrB,Multidrug efflux pump subunit AcrB | | Authors: | Sjuts, H, Ornik, A.R, Pos, K.M. | | Deposit date: | 2015-11-09 | | Release date: | 2016-04-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Molecular basis for inhibition of AcrB multidrug efflux pump by novel and powerful pyranopyridine derivatives.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5ENS
 
 | | Rhodamine bound structure of bacterial efflux pump. | | Descriptor: | DARPin, Multidrug efflux pump subunit AcrB,Multidrug efflux pump subunit AcrB, RHODAMINE 6G | | Authors: | Sjuts, H, Ornik, A.R, Pos, K.M. | | Deposit date: | 2015-11-09 | | Release date: | 2016-04-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Molecular basis for inhibition of AcrB multidrug efflux pump by novel and powerful pyranopyridine derivatives.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5EN5
 
 | | Apo structure of bacterial efflux pump. | | Descriptor: | DARPin, Multidrug efflux pump subunit AcrB,Multidrug efflux pump subunit AcrB | | Authors: | Sjuts, H, Ornik, A.R, Pos, K.M. | | Deposit date: | 2015-11-09 | | Release date: | 2016-04-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Molecular basis for inhibition of AcrB multidrug efflux pump by novel and powerful pyranopyridine derivatives.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
6JKK
 
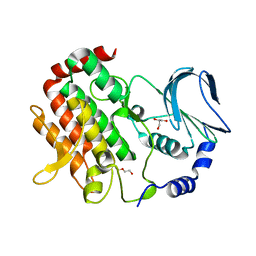 | | Crystal structure of BubR1 kinase domain | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, Mitotic checkpoint control protein kinase BUB1 | | Authors: | Lin, L, Ye, S, Huang, Y, Liu, X, Zhang, R, Yao, X. | | Deposit date: | 2019-03-01 | | Release date: | 2019-06-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | BubR1 phosphorylates CENP-E as a switch enabling the transition from lateral association to end-on capture of spindle microtubules.
Cell Res., 29, 2019
|
|
6JKM
 
 | | Crystal structure of BubR1 kinase domain | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Lin, L, Ye, S, Huang, Y, Liu, X, Zhang, R, Yao, X. | | Deposit date: | 2019-03-01 | | Release date: | 2019-06-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | BubR1 phosphorylates CENP-E as a switch enabling the transition from lateral association to end-on capture of spindle microtubules.
Cell Res., 29, 2019
|
|
5YC0
 
 | | Crystal structure of LP-46/N44 | | Descriptor: | Envelope glycoprotein, LP-46 | | Authors: | Zhang, X, Wang, X, He, Y. | | Deposit date: | 2017-09-05 | | Release date: | 2018-02-14 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Exceptional potency and structural basis of a T1249-derived lipopeptide fusion inhibitor against HIV-1, HIV-2, and simian immunodeficiency virus
J. Biol. Chem., 293, 2018
|
|
8HDJ
 
 | |
8HER
 
 | |
8HEP
 
 | |
8HEQ
 
 | |
7WNT
 
 | | RNase J from Mycobacterium tuberculosis | | Descriptor: | Ribonuclease J, ZINC ION | | Authors: | Li, J, Bao, L, Hu, J, Zhan, B, Li, Z. | | Deposit date: | 2022-01-19 | | Release date: | 2023-01-25 | | Last modified: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Structural insights into RNase J that plays an essential role in Mycobacterium tuberculosis RNA metabolism
Nat Commun, 14, 2023
|
|
7WNU
 
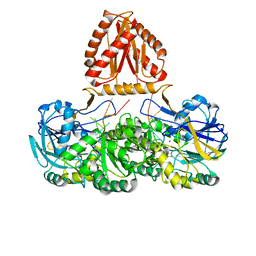 | | Mycobacterium tuberculosis Rnase J complex with 7nt RNA | | Descriptor: | RNA (5'-R(P*AP*AP*AP*AP*AP*AP*A)-3'), Ribonuclease J, ZINC ION | | Authors: | Li, J, Hu, J, Bao, L, Zhan, B, Li, Z. | | Deposit date: | 2022-01-19 | | Release date: | 2023-01-25 | | Last modified: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural insights into RNase J that plays an essential role in Mycobacterium tuberculosis RNA metabolism
Nat Commun, 14, 2023
|
|
5Z0W
 
 | |
6IVU
 
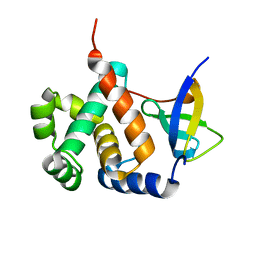 | |
6IVS
 
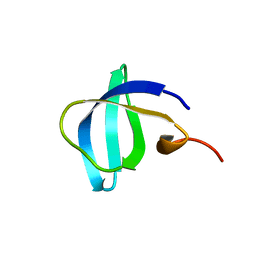 | |
6KTS
 
 | | Structure of C34N126K/N36 | | Descriptor: | Envelope glycoprotein, Glycoprotein 41 | | Authors: | Yu, D.W, Qin, B. | | Deposit date: | 2019-08-28 | | Release date: | 2020-09-16 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural and Functional Characterization of the Secondary Mutation N126K Selected by Various HIV-1 Fusion Inhibitors.
Viruses, 12, 2020
|
|
4MF1
 
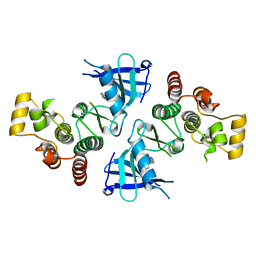 | | ITK kinase domain in complex with benzothiazole inhibitor 12b (1S,2S)-2-{4-[(DIMETHYLAMINO)METHYL]PHENYL}-N-[6-(1H-PYRAZOL-4-YL)-1,3-BENZOTHIAZOL-2-YL]CYCLOPROPANECARBOXAMIDE | | Descriptor: | (1S,2S)-2-{4-[(dimethylamino)methyl]phenyl}-N-[6-(1H-pyrazol-4-yl)-1,3-benzothiazol-2-yl]cyclopropanecarboxamide, Tyrosine-protein kinase ITK/TSK | | Authors: | Eigenbrot, C, Shia, S. | | Deposit date: | 2013-08-27 | | Release date: | 2013-11-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.113 Å) | | Cite: | Structure-based design and synthesis of potent benzothiazole inhibitors of interleukin-2 inducible T cell kinase (ITK).
Bioorg.Med.Chem.Lett., 23, 2013
|
|
4MF0
 
 | | ITK kinase domain in complex with benzothiazole inhibitor compound 12a (1S,2S)-2-{4-[(DIMETHYLAMINO)METHYL]PHENYL}-N-[6-(PYRIDIN-3-YL)-1,3-BENZOTHIAZOL-2-YL]CYCLOPROPANECARBOXAMIDE (12a) | | Descriptor: | (1S,2S)-2-{4-[(dimethylamino)methyl]phenyl}-N-[6-(pyridin-3-yl)-1,3-benzothiazol-2-yl]cyclopropanecarboxamide, Tyrosine-protein kinase ITK/TSK | | Authors: | Eigenbrot, C, Shia, S. | | Deposit date: | 2013-08-27 | | Release date: | 2013-11-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.67 Å) | | Cite: | Structure-based design and synthesis of potent benzothiazole inhibitors of interleukin-2 inducible T cell kinase (ITK).
Bioorg.Med.Chem.Lett., 23, 2013
|
|
1ELD
 
 | | Structural analysis of the active site of porcine pancreatic elastase based on the x-ray crystal structures of complexes with trifluoroacetyl-dipeptide-anilide inhibitors | | Descriptor: | ACETIC ACID, CALCIUM ION, ELASTASE, ... | | Authors: | Mattos, C, Petsko, G.A, Ringe, D. | | Deposit date: | 1994-10-24 | | Release date: | 1995-02-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural analysis of the active site of porcine pancreatic elastase based on the X-ray crystal structures of complexes with trifluoroacetyl-dipeptide-anilide inhibitors.
Biochemistry, 34, 1995
|
|
1ELE
 
 | | STRUCTURAL ANALYSIS OF THE ACTIVE SITE OF PORCINE PANCREATIC ELASTASE BASED ON THE X-RAY CRYSTAL STRUCTURES OF COMPLEXES WITH TRIFLUOROACETYL-DIPEPTIDE-ANILIDE INHIBITORS | | Descriptor: | CALCIUM ION, ELASTASE, N-(trifluoroacetyl)-L-valyl-N-[4-(trifluoromethyl)phenyl]-L-alaninamide, ... | | Authors: | Mattos, C, Petsko, G.A, Ringe, D. | | Deposit date: | 1994-10-24 | | Release date: | 1995-02-14 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural analysis of the active site of porcine pancreatic elastase based on the X-ray crystal structures of complexes with trifluoroacetyl-dipeptide-anilide inhibitors.
Biochemistry, 34, 1995
|
|
4PPB
 
 | |
4PPA
 
 | |
4PPC
 
 | |
4PP9
 
 | |
