8OG8
 
 | | Crystal structure of human DCAF1 WD40 repeats (Q1250L) in complex with compound 3 | | Descriptor: | 1,2-ETHANEDIOL, 5-(2-methyl-1-phenyl-propan-2-yl)imidazo[2,1-a]isoquinoline, ACETATE ION, ... | | Authors: | Schroeder, M, Vulpetti, A, Renatus, M. | | Deposit date: | 2023-03-19 | | Release date: | 2023-06-14 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Discovery of New Binders for DCAF1, an Emerging Ligase Target in the Targeted Protein Degradation Field.
Acs Med.Chem.Lett., 14, 2023
|
|
8OG6
 
 | | Crystal structure of human DCAF1 WD40 repeats (Q1250L) in complex with compound 1 | | Descriptor: | 5-(2-fluorophenyl)-2,3-dihydroimidazo[2,1-a]isoquinoline, ACETATE ION, DDB1- and CUL4-associated factor 1 | | Authors: | Schroeder, M, Vulpetti, A, Renatus, M. | | Deposit date: | 2023-03-19 | | Release date: | 2023-06-14 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.245 Å) | | Cite: | Discovery of New Binders for DCAF1, an Emerging Ligase Target in the Targeted Protein Degradation Field.
Acs Med.Chem.Lett., 14, 2023
|
|
8OG9
 
 | | Crystal structure of human DCAF1 WD40 repeats (Q1250L) in complex with compound 4 | | Descriptor: | 5-[1-(4-chlorophenyl)cyclopropyl]imidazo[2,1-a]isoquinoline, DDB1- and CUL4-associated factor 1 | | Authors: | Schroeder, M, Vulpetti, A, Renatus, M. | | Deposit date: | 2023-03-19 | | Release date: | 2023-06-14 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.945 Å) | | Cite: | Discovery of New Binders for DCAF1, an Emerging Ligase Target in the Targeted Protein Degradation Field.
Acs Med.Chem.Lett., 14, 2023
|
|
8OGB
 
 | | Crystal structure of human DCAF1 WD40 repeats (Q1250L) in complex with compound 8 | | Descriptor: | 1,2-ETHANEDIOL, DDB1- and CUL4-associated factor 1, DIMETHYL SULFOXIDE, ... | | Authors: | Schroeder, M, Vulpetti, A, Renatus, M. | | Deposit date: | 2023-03-19 | | Release date: | 2023-06-14 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Discovery of New Binders for DCAF1, an Emerging Ligase Target in the Targeted Protein Degradation Field.
Acs Med.Chem.Lett., 14, 2023
|
|
8OGC
 
 | | Crystal structure of human DCAF1 WD40 repeats (Q1250L) in complex with compound 11 | | Descriptor: | 1,2-ETHANEDIOL, 1-[4-[4-(2-azanylethylamino)-2-[1-(4-chlorophenyl)cyclopentyl]quinazolin-7-yl]piperazin-1-yl]ethanone, ACETATE ION, ... | | Authors: | Schroeder, M, Vulpetti, A, Renatus, M. | | Deposit date: | 2023-03-19 | | Release date: | 2023-06-14 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Discovery of New Binders for DCAF1, an Emerging Ligase Target in the Targeted Protein Degradation Field.
Acs Med.Chem.Lett., 14, 2023
|
|
8OG7
 
 | | Crystal structure of human DCAF1 WD40 repeats (Q1250L) in complex with compound 2 | | Descriptor: | (4~{R})-4-[3-(4-chloranylphenoxy)phenyl]pyrrolidin-2-imine, 1,2-ETHANEDIOL, ACETATE ION, ... | | Authors: | Schroeder, M, Vulpetti, A, Renatus, M. | | Deposit date: | 2023-03-19 | | Release date: | 2023-06-14 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Discovery of New Binders for DCAF1, an Emerging Ligase Target in the Targeted Protein Degradation Field.
Acs Med.Chem.Lett., 14, 2023
|
|
8OGA
 
 | | Crystal structure of human DCAF1 WD40 repeats (Q1250L) in complex with compound 6 | | Descriptor: | 1,2-ETHANEDIOL, 5-[1-(4-methoxyphenyl)cyclopropyl]-8-(4-methylpiperazin-1-yl)-2,3-dihydroimidazo[2,1-a]isoquinoline, DDB1- and CUL4-associated factor 1, ... | | Authors: | Schroeder, M, Vulpetti, A, Renatus, M. | | Deposit date: | 2023-03-19 | | Release date: | 2023-06-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Discovery of New Binders for DCAF1, an Emerging Ligase Target in the Targeted Protein Degradation Field.
Acs Med.Chem.Lett., 14, 2023
|
|
8P0M
 
 | | Crystal structure of TEAD3 in complex with IAG933 | | Descriptor: | 2-(2-{2-[2-(2-METHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHANOL, 4-[(2~{S})-5-chloranyl-6-fluoranyl-2-phenyl-2-[(2~{S})-pyrrolidin-2-yl]-3~{H}-1-benzofuran-4-yl]-5-fluoranyl-6-(2-hydroxyethyloxy)-~{N}-methyl-pyridine-3-carboxamide, DIMETHYL SULFOXIDE, ... | | Authors: | Scheufler, C, Villard, F, Chau, S. | | Deposit date: | 2023-05-10 | | Release date: | 2024-04-10 | | Last modified: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (1.961 Å) | | Cite: | Direct and selective pharmacological disruption of the YAP-TEAD interface by IAG933 inhibits Hippo-dependent and RAS-MAPK-altered cancers.
Nat Cancer, 5, 2024
|
|
6PFY
 
 | |
4UZD
 
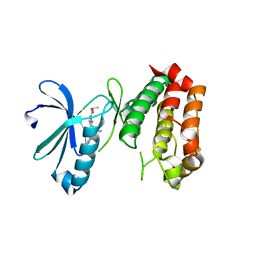 | | SAR156497 an exquisitely selective inhibitor of Aurora kinases | | Descriptor: | AURORA KINASE A, ethyl (9S)-9-[3-(1H-benzimidazol-2-yloxy)phenyl]-8-oxo-4,5,6,7,8,9-hexahydro-2H-pyrrolo[3,4-b]quinoline-3-carboxylate | | Authors: | Pouzieux, S, Delarbre, L, Crenne, J.Y. | | Deposit date: | 2014-09-05 | | Release date: | 2014-11-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Sar156497 an Exquisitely Selective Inhibitor of Aurora Kinases.
J.Med.Chem., 58, 2015
|
|
4UZH
 
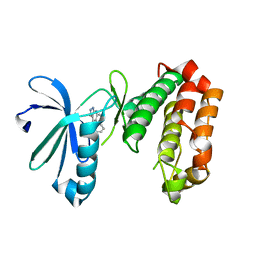 | | SAR156497 an exquisitely selective inhibitor of Aurora kinases | | Descriptor: | (4S)-4-(2-fluorophenyl)-2,4,6,7,8,9-hexahydro-5H-pyrazolo[3,4-b][1,7]naphthyridin-5-one, AURORA 2 KINASE DOMAIN | | Authors: | Pouzieux, S, Maignan, S, Crenne, J.Y. | | Deposit date: | 2014-09-05 | | Release date: | 2014-11-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Sar156497 an Exquisitely Selective Inhibitor of Aurora Kinases.
J.Med.Chem., 58, 2015
|
|
8S3X
 
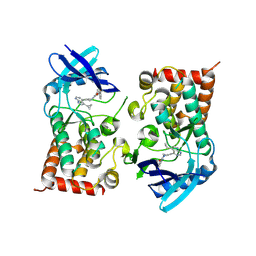 | | LIM Domain Kinase 2 (LIMK2) bound to compound 52 | | Descriptor: | 4-(5-cyclopropyl-7~{H}-pyrrolo[2,3-d]pyrimidin-4-yl)-~{N}-[3-(3-methoxyphenyl)phenyl]-3,6-dihydro-2~{H}-pyridine-1-carboxamide, LIM domain kinase 2 | | Authors: | Mathea, S, Chatterjee, D, Preuss, F, Ple, K, Knapp, S. | | Deposit date: | 2024-02-20 | | Release date: | 2024-03-06 | | Last modified: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Tetrahydropyridine LIMK inhibitors: Structure activity studies and biological characterization.
Eur.J.Med.Chem., 271, 2024
|
|
5J5D
 
 | |
7OPS
 
 | | Crystal structure of haspin in complex with ZW282 (compound 2a) | | Descriptor: | 2-methylsulfanyl-10-nitro-pyrido[3,4-g]quinazoline, GLYCEROL, Serine/threonine-protein kinase haspin | | Authors: | Chaikuad, A, Anizon, F, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2021-06-01 | | Release date: | 2021-07-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Synthesis and biological evaluation of Haspin inhibitors: Kinase inhibitory potency and cellular activity.
Eur.J.Med.Chem., 236, 2022
|
|
2MD0
 
 | | Solution structure of ShK-like immunomodulatory peptide from Ancylostoma caninum (hookworm) | | Descriptor: | AcK1 | | Authors: | Chhabra, S, Swarbrick, J.D, Mohanty, B, Chang, S.C, Chandy, G.K, Pennington, M.W, Norton, R.S. | | Deposit date: | 2013-08-28 | | Release date: | 2014-07-02 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Kv1.3 channel-blocking immunomodulatory peptides from parasitic worms: implications for autoimmune diseases.
Faseb J., 28, 2014
|
|
1CEC
 
 | |
1XYZ
 
 | |
2MCR
 
 | | Solution structure of ShK-like immunomodulatory peptide from Brugia malayi (filarial worm) | | Descriptor: | Probable zinc metalloproteinase, putative | | Authors: | Chhabra, S, Swarbrick, J.D, Pennington, M.W, Chang, S.C, Norton, R.S. | | Deposit date: | 2013-08-22 | | Release date: | 2014-07-02 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Kv1.3 channel-blocking immunomodulatory peptides from parasitic worms: implications for autoimmune diseases.
Faseb J., 28, 2014
|
|
6CO1
 
 | |
6CO2
 
 | | Structure of an engineered protein (NUDT16TI) in complex with 53BP1 Tudor domains | | Descriptor: | NUDT16-Tudor-interacting (NUDT16TI), TP53-binding protein 1 | | Authors: | Botuyan, M.V, Thompson, J.R, Cui, G, Mer, G. | | Deposit date: | 2018-03-10 | | Release date: | 2018-06-06 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Mechanism of 53BP1 activity regulation by RNA-binding TIRR and a designer protein.
Nat. Struct. Mol. Biol., 25, 2018
|
|
6D0L
 
 | | Structure of human TIRR | | Descriptor: | Tudor-interacting repair regulator protein | | Authors: | Cui, G, Botuyan, M.V, Mer, G. | | Deposit date: | 2018-04-10 | | Release date: | 2018-06-06 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Mechanism of 53BP1 activity regulation by RNA-binding TIRR and a designer protein.
Nat. Struct. Mol. Biol., 25, 2018
|
|
2K9E
 
 | |
5E0N
 
 | | Crystal Structure of MSMEG_3139, a monofunctional enoyl CoA isomerase from M.smegmatis | | Descriptor: | Enoyl-CoA hydratase/isomerase | | Authors: | Priyadarshan, K, Haque, A.S, Anandakrishnan, M, Sankaranarayanan, R. | | Deposit date: | 2015-09-29 | | Release date: | 2016-02-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.061 Å) | | Cite: | Unsaturated Lipid Assimilation by Mycobacteria Requires Auxiliary cis-trans Enoyl CoA Isomerase.
Chem.Biol., 22, 2015
|
|
5WCV
 
 | |
5H3L
 
 | | Structure of methylglyoxal synthase crystallised as a contaminant | | Descriptor: | FORMIC ACID, Methylglyoxal synthase | | Authors: | Hatti, K, Dadireddy, V, Srinivasan, N, Ramakumar, S, Murthy, M.R.N. | | Deposit date: | 2016-10-25 | | Release date: | 2016-11-09 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure determination of contaminant proteins using the MarathonMR procedure.
J. Struct. Biol., 197, 2017
|
|
