8VS5
 
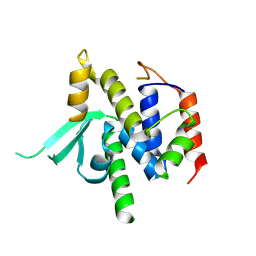 | | Structure of catalytic domain of telomere resolvase, ResT, from Borrelia garinii | | Descriptor: | Telomere resolvase ResT | | Authors: | Semper, C, Savchenko, A, Watanabe, N, Center for Structural Biology of Infectious Diseases (CSBID) | | Deposit date: | 2024-01-23 | | Release date: | 2024-07-17 | | Last modified: | 2024-08-28 | | Method: | X-RAY DIFFRACTION (2.796 Å) | | Cite: | Structure analysis of the telomere resolvase from the Lyme disease spirochete Borrelia garinii reveals functional divergence of its C-terminal domain.
Nucleic Acids Res., 52, 2024
|
|
8VJ1
 
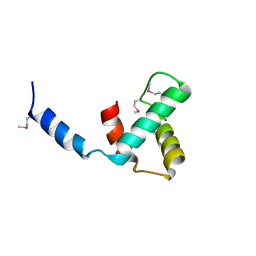 | | Structure of C-terminal domain of telomere resolvase, ResT, from Borrelia garinii | | Descriptor: | Telomere resolvase ResT | | Authors: | Semper, C, Savchenko, A, Watanabe, N, Center for Structural Biology of Infectious Diseases (CSBID) | | Deposit date: | 2024-01-05 | | Release date: | 2024-07-17 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Structure analysis of the telomere resolvase from the Lyme disease spirochete Borrelia garinii reveals functional divergence of its C-terminal domain.
Nucleic Acids Res., 52, 2024
|
|
8VDX
 
 | | Crystal structure of bacterial extracellular solute-binding protein from Bordetella bronchiseptica RB50 | | Descriptor: | ACETATE ION, GLYCEROL, TETRAETHYLENE GLYCOL, ... | | Authors: | Chang, C, Tesar, C, Endres, M, Joachimiak, A, Center for Structural Biology of Infectious Diseases (CSBID) | | Deposit date: | 2023-12-18 | | Release date: | 2024-12-18 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structure of bacterial extracellular solute-binding protein from Bordetella bronchiseptica RB50
To Be Published
|
|
8VLW
 
 | |
8F4Y
 
 | | Crystal Structure of SARS-CoV-2 2'-O-Methyltransferase in Complex with Compound 5a covalently bound to nsp16 and nsp10 | | Descriptor: | 2'-O-methyltransferase, 4-[(E)-2-(2,4-dichlorophenyl)ethenyl]-6-(trifluoromethyl)pyrimidin-2-ol, 4-[2-(2,4-dichlorophenyl)ethyl]-6-(trifluoromethyl)pyrimidin-2-ol, ... | | Authors: | Minasov, G, Shuvalova, L, Brunzelle, J.S, Rosas-Lemus, M, Kiryukhina, O, Satchell, K.J.F, Center for Structural Biology of Infectious Diseases (CSBID) | | Deposit date: | 2022-11-11 | | Release date: | 2023-10-18 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Discovery of a Druggable, Cryptic Pocket in SARS-CoV-2 nsp16 Using Allosteric Inhibitors.
Acs Infect Dis., 9, 2023
|
|
8F4S
 
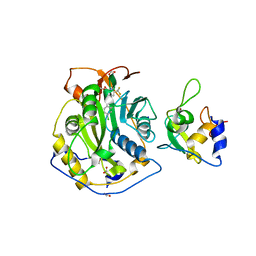 | | Crystal Structure of the SARS-CoV-2 2'-O-Methyltransferase with Compound 5a bound to the Cryptic Pocket of nsp16 | | Descriptor: | 2'-O-methyltransferase, 4-[(E)-2-(2,4-dichlorophenyl)ethenyl]-6-(trifluoromethyl)pyrimidin-2-ol, FORMIC ACID, ... | | Authors: | Minasov, G, Shuvalova, L, Brunzelle, J.S, Rosas-Lemus, M, Kiryukhina, O, Satchell, K.J.F, Center for Structural Biology of Infectious Diseases (CSBID) | | Deposit date: | 2022-11-11 | | Release date: | 2023-10-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Discovery of a Druggable, Cryptic Pocket in SARS-CoV-2 nsp16 Using Allosteric Inhibitors.
Acs Infect Dis., 9, 2023
|
|
8EBC
 
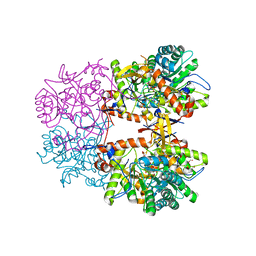 | | Crystal Structure of the Catalytic Domain of the Inosine Monophosphate Dehydrogenase from Listeria monocytogenes in the complex with IMP | | Descriptor: | FORMIC ACID, GLYCEROL, INOSINIC ACID, ... | | Authors: | Kim, Y, Maltseva, N, Makowska-Grzyska, M, Osipiuk, J, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID), Center for Structural Biology of Infectious Diseases (CSBID) | | Deposit date: | 2022-08-31 | | Release date: | 2022-09-07 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of the Catalytic Domain of the Inosine Monophosphate Dehydrogenase from Listeria monocytogenes in the complex with IMP
To Be Published
|
|
8EP7
 
 | | Crystal Structure of the Ketol-acid Reductoisomerase from Bacillus anthracis in complex with NADP | | Descriptor: | ACETIC ACID, Ketol-acid reductoisomerase (NADP(+)) 2, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Kim, Y, Maltseva, N, Osipiuk, J, Gu, M, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID), Center for Structural Biology of Infectious Diseases (CSBID) | | Deposit date: | 2022-10-05 | | Release date: | 2022-10-19 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of the Ketol-acid Reductoisomerase from Bacillus anthracis in the complex with NADP.
To Be Published
|
|
8EP6
 
 | | Crystal Structure of the Beta-lactamase Class D from Chitinophaga pinensis in complex with Avibactam | | Descriptor: | (2S,5R)-1-formyl-5-[(sulfooxy)amino]piperidine-2-carboxamide, ACETIC ACID, Beta-lactamase Class D Cpin_0907 | | Authors: | Maltseva, N, Kim, Y, Endres, M, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID), Center for Structural Biology of Infectious Diseases (CSBID) | | Deposit date: | 2022-10-05 | | Release date: | 2022-10-19 | | Last modified: | 2023-02-01 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal Structure of the Beta-lactamase Class D from Chitinophaga pinensis in the complex with Avibactam.
To Be Published
|
|
8EW7
 
 | | Human Serum Albumin with Cobalt (II) and Myristic Acid - crystal 2 | | Descriptor: | COBALT (II) ION, MYRISTIC ACID, Serum albumin | | Authors: | Gucwa, M, Cooper, D.R, Stewart, A.J, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID), Center for Structural Biology of Infectious Diseases (CSBID) | | Deposit date: | 2022-10-21 | | Release date: | 2022-11-09 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural and biochemical characterisation of Co2+-binding sites on serum albumins and their interplay with fatty acids
Chem Sci, 14, 2023
|
|
8EW4
 
 | | Human Serum Albumin with Cobalt (II) and Myristic Acid - crystal 1 | | Descriptor: | COBALT (II) ION, MYRISTIC ACID, Serum albumin | | Authors: | Gucwa, M, Cooper, D.R, Unciano, J, Lea, K, Kim, L, Lenkiewicz, J, Starban, I, Stewart, A.J, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID), Center for Structural Biology of Infectious Diseases (CSBID) | | Deposit date: | 2022-10-21 | | Release date: | 2022-11-09 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural and biochemical characterisation of Co2+-binding sites on serum albumins and their interplay with fatty acids
Chem Sci, 14, 2023
|
|
8EY5
 
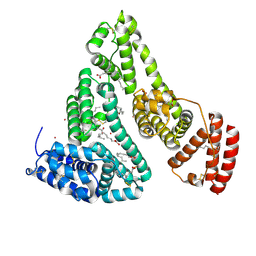 | | Human Serum Albumin with Cobalt (II) and Myristic Acid - crystal 3 | | Descriptor: | COBALT (II) ION, MYRISTIC ACID, Serum albumin | | Authors: | Gucwa, M, Cooper, D.R, Stewart, A.J, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID), Center for Structural Biology of Infectious Diseases (CSBID) | | Deposit date: | 2022-10-26 | | Release date: | 2022-11-09 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural and biochemical characterisation of Co2+-binding sites on serum albumins and their interplay with fatty acids
Chem Sci, 14, 2023
|
|
8EWO
 
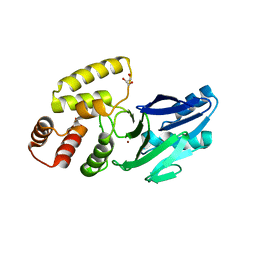 | | Crystal structure of putative glyoxylase II from Pseudomonas aeruginosa | | Descriptor: | CHLORIDE ION, GLYCEROL, PA1813, ... | | Authors: | Stogios, P.J, Skarina, T, Endres, M, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID), Center for Structural Biology of Infectious Diseases (CSBID) | | Deposit date: | 2022-10-24 | | Release date: | 2022-11-16 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Crystal structure of putative glyoxylase II from Pseudomonas aeruginosa
To Be Published
|
|
8F09
 
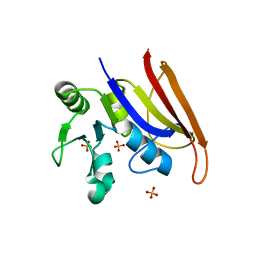 | | Crystal structure of a trimethoprim-resistant dihydrofolate reductase (DHFR) enzyme from an uncultured soil bacterium | | Descriptor: | Dihydrofolate reductase, SULFATE ION | | Authors: | Stogios, P.J, Evdokimova, D, Borek, D, Di Leo, R, Semper, C, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Biology of Infectious Diseases (CSBID) | | Deposit date: | 2022-11-02 | | Release date: | 2022-11-16 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Crystal structure of a trimethoprim-resistant dihydrofolate reductase (DHFR) enzyme from an uncultured soil bacterium
To Be Published
|
|
9DRS
 
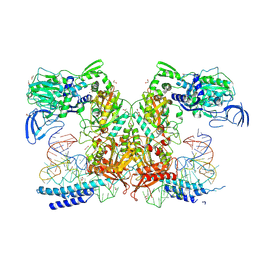 | | Crystal structure of M. tuberculosis PheRS-tRNA complex bound to inhibitor D-116 | | Descriptor: | 1,2-ETHANEDIOL, 1H-benzimidazol-2-amine, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Gade, P, Chang, C, Forte, B, Wower, J, Gilbert, I.H, Baragana, B, Michalska, K, Joachimiak, A, Center for Structural Biology of Infectious Diseases (CSBID) | | Deposit date: | 2024-09-26 | | Release date: | 2025-02-12 | | Last modified: | 2025-02-19 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Different chemical scaffolds bind to L-phe site in Mycobacterium tuberculosis Phe-tRNA synthetase.
Eur.J.Med.Chem., 287, 2025
|
|
9DRV
 
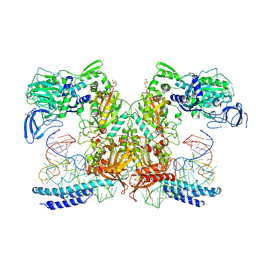 | | Crystal structure of M. tuberculosis PheRS-tRNA complex bound to inhibitor D-004 | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ACETATE ION, ... | | Authors: | Gade, P, Chang, C, Forte, B, Wower, J, Gilbert, I.H, Baragana, B, Michalska, K, Joachimiak, A, Center for Structural Biology of Infectious Diseases (CSBID) | | Deposit date: | 2024-09-26 | | Release date: | 2025-02-12 | | Last modified: | 2025-02-19 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Different chemical scaffolds bind to L-phe site in Mycobacterium tuberculosis Phe-tRNA synthetase.
Eur.J.Med.Chem., 287, 2025
|
|
9DRT
 
 | | Crystal structure of the complex of M. tuberculosis PheRS with cognate precursor tRNA and fragment DDD00805735 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ACETATE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Chang, C, Michalska, K, Forte, B, Baragana, B, Gilbert, I.H, Wower, J, Joachimiak, A, Center for Structural Biology of Infectious Diseases (CSBID) | | Deposit date: | 2024-09-26 | | Release date: | 2025-06-04 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Different chemical scaffolds bind to L-phe site in Mycobacterium tuberculosis Phe-tRNA synthetase.
Eur.J.Med.Chem., 287, 2025
|
|
9EDV
 
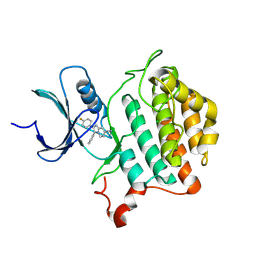 | | Crystal structure of Yck2 from Candida albicans in complex with inhibitor 1e: 2-(4-fluorophenyl)-3-(pyridin-4-yl)imidazo[1,2-a]pyridine-7-carbonitrile | | Descriptor: | 2-(4-fluorophenyl)-3-(pyridin-4-yl)imidazo[1,2-a]pyridine-7-carbonitrile, CHLORIDE ION, non-specific serine/threonine protein kinase | | Authors: | Stogios, P.J, Whitesell, L, Cowen, L.E, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Biology of Infectious Diseases (CSBID) | | Deposit date: | 2024-11-18 | | Release date: | 2025-01-08 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Crystal structure of Yck2 from Candida albicans in complex with inhibitor 1e: 2-(4-fluorophenyl)-3-(pyridin-4-yl)imidazo[1,2-a]pyridine-7-carbonitrile
To Be Published
|
|
9EDX
 
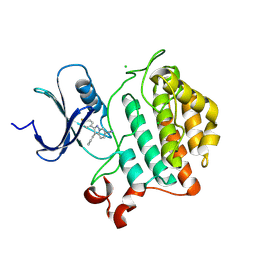 | | Crystal structure of Yck2 from Candida albicans in complex with inhibitor 2a: 2-(4-fluorophenyl)-3-(pyridin-4-yl)pyrazolo[1,5-a]pyridine-6-carbonitrile | | Descriptor: | 2-(4-fluorophenyl)-3-(pyridin-4-yl)pyrazolo[1,5-a]pyridine-6-carbonitrile, CHLORIDE ION, non-specific serine/threonine protein kinase | | Authors: | Stogios, P.J, Whitesell, L, Cowen, L.E, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Biology of Infectious Diseases (CSBID) | | Deposit date: | 2024-11-18 | | Release date: | 2025-01-08 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Crystal structure of Yck2 from Candida albicans in complex with inhibitor 2a: 2-(4-fluorophenyl)-3-(pyridin-4-yl)pyrazolo[1,5-a]pyridine-6-carbonitrile
To Be Published
|
|
9EDY
 
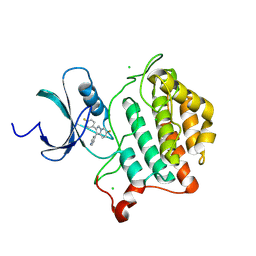 | | Crystal structure of Yck2 from Candida albicans in complex with inhibitor 2b: 6-fluoro-2-(4-fluorophenyl)-3-(pyridin-4-yl)pyrazolo[1,5-a]pyridine | | Descriptor: | 6-fluoro-2-(4-fluorophenyl)-3-(pyridin-4-yl)pyrazolo[1,5-a]pyridine, CHLORIDE ION, non-specific serine/threonine protein kinase | | Authors: | Stogios, P.J, Whitesell, L, Cowen, L.E, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Biology of Infectious Diseases (CSBID) | | Deposit date: | 2024-11-18 | | Release date: | 2025-01-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of Yck2 from Candida albicans in complex with inhibitor 2b: 6-fluoro-2-(4-fluorophenyl)-3-(pyridin-4-yl)pyrazolo[1,5-a]pyridine
To Be Published
|
|
9EDW
 
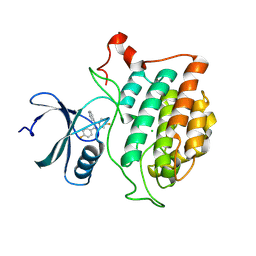 | | Crystal structure of Yck2 from Candida albicans in complex with inhibitor 1f: 7-fluoro-2-(4-fluorophenyl)-3-(pyridin-4-yl)imidazo[1,2-a]pyridine | | Descriptor: | 7-fluoro-2-(4-fluorophenyl)-3-(pyridin-4-yl)imidazo[1,2-a]pyridine, CHLORIDE ION, Non-specific serine/threonine protein kinase | | Authors: | Stogios, P.J, Whitesell, L, Cowen, L.E, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Biology of Infectious Diseases (CSBID) | | Deposit date: | 2024-11-18 | | Release date: | 2025-01-08 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Crystal structure of Yck2 from Candida albicans in complex with inhibitor 1f: 7-fluoro-2-(4-fluorophenyl)-3-(pyridin-4-yl)imidazo[1,2-a]pyridine
To Be Published
|
|
8SQN
 
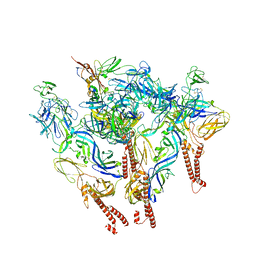 | | CryoEM structure of Western equine encephalitis virus VLP in complex with the chimeric Du-D1-Mo-D2 MXRA8 receptor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, DuD1MoD2 chimeric MXRA8, E1 envelope glycoprotein, ... | | Authors: | Zimmerman, M.I, Fremont, D.H, Center for Structural Genomics of Infectious Diseases (CSGID), Center for Structural Biology of Infectious Diseases (CSBID) | | Deposit date: | 2023-05-04 | | Release date: | 2023-12-20 | | Last modified: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (3.89 Å) | | Cite: | Vertebrate-class-specific binding modes of the alphavirus receptor MXRA8.
Cell, 186, 2023
|
|
9AVI
 
 | |
9BHE
 
 | |
9BHC
 
 | |
