7UZP
 
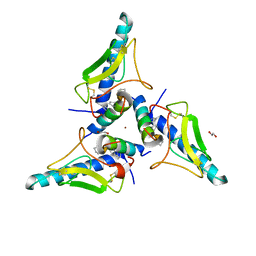 | | parathyroid hormone 1 receptor extracellular domain complexed with a peptide ligand containing three beta-amino acids | | Descriptor: | 1,2-ETHANEDIOL, PTHrP[1-36] 24,28,31 XCP, Parathyroid hormone/parathyroid hormone-related peptide receptor, ... | | Authors: | Yu, Z, Bruchs, A.T, Bingman, C.A, Gellman, S.H. | | Deposit date: | 2022-05-09 | | Release date: | 2022-10-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Altered signaling at the PTH receptor via modified agonist contacts with the extracellular domain provides a path to prolonged agonism in vivo.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
5F4Z
 
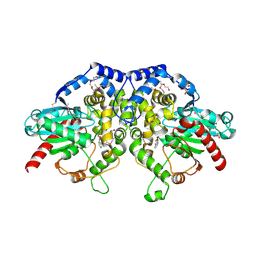 | | The crystal structure of an epoxide hydrolase from Streptomyces carzinostaticus subsp. neocarzinostaticus | | Descriptor: | (1~{R},2~{R})-2,3-dihydro-1~{H}-indene-1,2-diol, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETATE ION, ... | | Authors: | Tan, K, Li, H, Jedrzejczak, R, BABNIGG, G, BINGMAN, C.A, YENNAMALLI, R, LOHMAN, J, Chang, C.Y, Shen, B, Phillips Jr, G.N, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2015-12-03 | | Release date: | 2016-02-17 | | Last modified: | 2020-09-23 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | The crystal structure of an epoxide hydrolase from Streptomyces carzinostaticus subsp. neocarzinostaticus
To Be Published
|
|
4FE3
 
 | |
7TA6
 
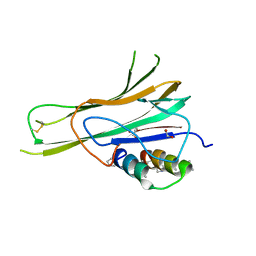 | | Trimer-to-Monomer Disruption of Tumor Necrosis Factor-alpha (TNF-alpha) by unnatural alpha/beta-peptide-1 | | Descriptor: | 1,2-ETHANEDIOL, AMINO GROUP, Alpha/Beta-peptide-1, ... | | Authors: | Niu, J, Bingman, C.A, Gellman, S.H. | | Deposit date: | 2021-12-20 | | Release date: | 2022-06-29 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.67 Å) | | Cite: | Trimer-to-Monomer Disruption Mechanism for a Potent, Protease-Resistant Antagonist of Tumor Necrosis Factor-alpha Signaling.
J.Am.Chem.Soc., 144, 2022
|
|
7TA3
 
 | |
6NIV
 
 | | Racemic Phenol-Soluble Modulin Alpha 3 Peptide | | Descriptor: | Phenol-soluble modulin PSM-alpha-3 | | Authors: | Yao, Z, Cary, B.P, Bingman, C.A, Wang, C, Kreitler, D.F, Satyshur, K.A, Forest, K.T, Gellman, S.H. | | Deposit date: | 2018-12-31 | | Release date: | 2019-05-15 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Use of a Stereochemical Strategy To Probe the Mechanism of Phenol-Soluble Modulin alpha 3 Toxicity.
J.Am.Chem.Soc., 141, 2019
|
|
6UI3
 
 | | GH5-4 broad specificity endoglucanase from Clostridum cellulovorans | | Descriptor: | 1,2-ETHANEDIOL, Cellulase | | Authors: | Bianchetti, C.M, Bingman, C.A, Smith, R.W, Glasgow, E.M, Fox, B.G. | | Deposit date: | 2019-09-30 | | Release date: | 2020-10-07 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | A structural and kinetic survey of GH5_4 endoglucanases reveals determinants of broad substrate specificity and opportunities for biomass hydrolysis.
J.Biol.Chem., 295, 2020
|
|
6WQV
 
 | | GH5-4 broad specificity endoglucanase from Ruminococcus champanellensis with bound cellotriose | | Descriptor: | 1,2-ETHANEDIOL, Endoglucanase, NITRATE ION, ... | | Authors: | Bianchetti, C.M, Bingman, C.A, Smith, R.W, Glasgow, E.M, Fox, B.G. | | Deposit date: | 2020-04-29 | | Release date: | 2020-11-18 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | A structural and kinetic survey of GH5_4 endoglucanases reveals determinants of broad substrate specificity and opportunities for biomass hydrolysis.
J.Biol.Chem., 295, 2020
|
|
3TOS
 
 | | Crystal Structure of CalS11, Calicheamicin Methyltransferase | | Descriptor: | 1,2-ETHANEDIOL, CalS11, GLUTAMIC ACID, ... | | Authors: | Chang, A, Aceti, D.J, Beebe, E.T, Makino, S.-I, Wrobel, R.L, Bingman, C.A, Thorson, J.S, Phillips Jr, G.N, Center for Eukaryotic Structural Genomics (CESG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2011-09-06 | | Release date: | 2011-10-05 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal Structure of CalS11, Calicheamicin methyltransferase
To be Published
|
|
3RSC
 
 | | Crystal Structure of CalG2, Calicheamicin Glycosyltransferase, TDP and calicheamicin T0 bound form | | Descriptor: | CalG2, Calicheamicin T0, PHOSPHATE ION, ... | | Authors: | Chang, A, Helmich, K.E, Singh, S, Bingman, C.A, Thorson, J.S, Phillips Jr, G.N, Center for Eukaryotic Structural Genomics (CESG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2011-05-02 | | Release date: | 2011-08-10 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Complete set of glycosyltransferase structures in the calicheamicin biosynthetic pathway reveals the origin of regiospecificity.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
5COW
 
 | | C. remanei PGL-1 Dimerization Domain | | Descriptor: | 1,2-ETHANEDIOL, Putative uncharacterized protein, SULFATE ION | | Authors: | Aoki, S.T, Bingman, C.A, Wickens, M, Kimble, J.E. | | Deposit date: | 2015-07-20 | | Release date: | 2016-02-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | PGL germ granule assembly protein is a base-specific, single-stranded RNase.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
1D78
 
 | | HIGH RESOLUTION REFINEMENT OF THE HEXAGONAL A-DNA OCTAMER D(GTGTACAC) AT 1.4 ANGSTROMS RESOLUTION | | Descriptor: | DNA (5'-D(*GP*TP*GP*TP*AP*CP*AP*C)-3') | | Authors: | Thota, N, Li, X.H, Bingman, C.A, Sundaralingam, M. | | Deposit date: | 1992-06-12 | | Release date: | 1993-04-15 | | Last modified: | 2023-03-22 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | High-resolution refinement of the hexagonal A-DNA octamer d(GTGTACAC) at 1.4 A.
Acta Crystallogr.,Sect.D, 49, 1993
|
|
5CV3
 
 | | C. remanei PGL-1 Dimerization Domain - Hg | | Descriptor: | ETHYL MERCURY ION, Putative uncharacterized protein | | Authors: | Aoki, S.T, Bingman, C.A, Wickens, M, Kimble, J.E. | | Deposit date: | 2015-07-25 | | Release date: | 2016-02-03 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3.17014766 Å) | | Cite: | PGL germ granule assembly protein is a base-specific, single-stranded RNase.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5CV1
 
 | | C. elegans PGL-1 Dimerization Domain | | Descriptor: | P granule abnormality protein 1 | | Authors: | Aoki, S.T, Bingman, C.A, Wickens, M, Kimble, J.E. | | Deposit date: | 2015-07-25 | | Release date: | 2016-02-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.599 Å) | | Cite: | PGL germ granule assembly protein is a base-specific, single-stranded RNase.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5DJE
 
 | | Crystal structure of the zuotin homology domain (ZHD) from yeast Zuo1 | | Descriptor: | 1,2-ETHANEDIOL, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Shrestha, O.K, Bingman, C.A, Craig, E.A. | | Deposit date: | 2015-09-02 | | Release date: | 2016-09-28 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Dual interaction of the Hsp70 J-protein cochaperone Zuotin with the 40S and 60S ribosomal subunits.
Nat.Struct.Mol.Biol., 23, 2016
|
|
6OWD
 
 | |
6O4M
 
 | | Racemic melittin | | Descriptor: | D-Melittin, Melittin, SULFATE ION | | Authors: | Kurgan, K.W, Bingman, C.A, Gellman, S.H, Forest, K.T. | | Deposit date: | 2019-02-28 | | Release date: | 2019-05-22 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.27 Å) | | Cite: | Retention of Native Quaternary Structure in Racemic Melittin Crystals.
J.Am.Chem.Soc., 141, 2019
|
|
6Q1I
 
 | |
6PZ7
 
 | | GH5-4 broad specificity endoglucanase from Clostridium acetobutylicum | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Endoglucanase family 5 | | Authors: | Bianchetti, C.M, Bingman, C.A, Fox, B.G. | | Deposit date: | 2019-07-31 | | Release date: | 2020-08-05 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | A structural and kinetic survey of GH5_4 endoglucanases reveals determinants of broad substrate specificity and opportunities for biomass hydrolysis.
J.Biol.Chem., 295, 2020
|
|
1D79
 
 | | HIGH RESOLUTION REFINEMENT OF THE HEXAGONAL A-DNA OCTAMER D(GTGTACAC) AT 1.4 ANGSTROMS RESOLUTION | | Descriptor: | DNA (5'-D(*GP*TP*GP*TP*AP*CP*AP*C)-3') | | Authors: | Thota, N, Li, X.H, Bingman, C.A, Sundaralingam, M. | | Deposit date: | 1992-06-12 | | Release date: | 1993-04-15 | | Last modified: | 2023-03-22 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | High-resolution refinement of the hexagonal A-DNA octamer d(GTGTACAC) at 1.4 A.
Acta Crystallogr.,Sect.D, 49, 1993
|
|
1Q45
 
 | | 12-0xo-phytodienoate reductase isoform 3 | | Descriptor: | 12-oxophytodienoate-10,11-reductase, FLAVIN MONONUCLEOTIDE | | Authors: | Phillips Jr, G.N, Johnson, K.A, Bingman, C.A, Smith, D.W, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2003-08-01 | | Release date: | 2003-11-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-ray structure of Arabidopsis At2g06050, 12-oxophytodienoate reductase isoform 3
Proteins, 58, 2005
|
|
1Q44
 
 | | Crystal Structure of an Arabidopsis Thaliana Putative Steroid Sulfotransferase | | Descriptor: | MALONIC ACID, Steroid Sulfotransferase | | Authors: | Phillips Jr, G.N, Smith, D.W, Johnson, K.A, Bingman, C.A, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2003-08-01 | | Release date: | 2003-11-25 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of At2g03760, a putative steroid sulfotransferase from Arabidopsis thaliana
Proteins, 57, 2004
|
|
4W79
 
 | | Crystal Structure of Human Protein N-terminal Glutamine Amidohydrolase | | Descriptor: | 1,2-ETHANEDIOL, CARBONATE ION, Protein N-terminal glutamine amidohydrolase, ... | | Authors: | Bitto, E, Bingman, C.A, McCoy, J.G, Wesenberg, G.E, Phillips Jr, G.N, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2014-08-21 | | Release date: | 2014-09-17 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of human protein N-terminal glutamine amidohydrolase, an initial component of the N-end rule pathway.
Plos One, 9, 2014
|
|
3H7C
 
 | | Crystal Structure of Arabidopsis thaliana Agmatine Deiminase from Cell Free Expression | | Descriptor: | 2,2',2''-NITRILOTRIETHANOL, Agmatine deiminase, CHLORIDE ION, ... | | Authors: | Burgie, E.S, Bingman, C.A, Phillips Jr, G.N, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2009-04-24 | | Release date: | 2009-05-26 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural Insights into the Catalytic Mechanism of Arabidopsis thaliana Agmatine Deiminase
To be Published
|
|
3IAA
 
 | | Crystal Structure of CalG2, Calicheamicin Glycosyltransferase, TDP bound form | | Descriptor: | CalG2, THYMIDINE-5'-DIPHOSPHATE | | Authors: | Chang, A, Singh, S, Bingman, C.A, Thorson, J.S, Phillips Jr, G.N. | | Deposit date: | 2009-07-13 | | Release date: | 2010-06-02 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (2.505 Å) | | Cite: | Complete set of glycosyltransferase structures in the calicheamicin biosynthetic pathway reveals the origin of regiospecificity.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
