2HNL
 
 | | Structure of the prostaglandin D synthase from the parasitic nematode Onchocerca volvulus | | Descriptor: | GLUTATHIONE, Glutathione S-transferase 1 | | Authors: | Perbandt, M, Hoppner, J, Betzel, C, Liebau, E. | | Deposit date: | 2006-07-13 | | Release date: | 2007-07-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the extracellular glutathione S-transferase OvGST1 from the human pathogenic parasite Onchocerca volvulus.
J.Mol.Biol., 377, 2008
|
|
2F6C
 
 | | Reaction geometry and thermostability of pyranose 2-oxidase from the white-rot fungus Peniophora sp., Thermostability mutant E542K | | Descriptor: | DI(HYDROXYETHYL)ETHER, FLAVIN-ADENINE DINUCLEOTIDE, Pyranose 2-oxidase, ... | | Authors: | Bannwarth, M, Heckmann-Pohl, D.M, Bastian, S, Giffhorn, F, Schulz, G.E. | | Deposit date: | 2005-11-29 | | Release date: | 2006-06-13 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Reaction Geometry and Thermostable Variant of Pyranose 2-Oxidase from the White-Rot Fungus Peniophora sp.
Biochemistry, 45, 2006
|
|
2F5V
 
 | | Reaction geometry and thermostability mutant of pyranose 2-oxidase from the white-rot fungus Peniophora sp. | | Descriptor: | DI(HYDROXYETHYL)ETHER, FLAVIN-ADENINE DINUCLEOTIDE, Pyranose 2-oxidase, ... | | Authors: | Bannwarth, M, Bastian, S, Heckmann-Pohl, D, Giffhorn, F, Schulz, G.E. | | Deposit date: | 2005-11-28 | | Release date: | 2006-06-13 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | Reaction Geometry and Thermostable Variant of Pyranose 2-Oxidase from the White-Rot Fungus Peniophora sp.
Biochemistry, 45, 2006
|
|
8RC1
 
 | |
3KZY
 
 | | Crystal structure of SNAP-tag | | Descriptor: | Methylated-DNA--protein-cysteine methyltransferase, ZINC ION | | Authors: | Bannwarth, M, Schmitt, S, Pojer, F, Schiltz, M, Johnsson, K. | | Deposit date: | 2009-12-09 | | Release date: | 2010-12-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | SNAP-tag structure
To be Published
|
|
7S7L
 
 | | Complex of tissue inhibitor of metalloproteinases-1 (TIMP-1) mutant (L34G/M66S/E67Y/L133N/S155L) with matrix metalloproteinase-3 catalytic domain (MMP-3cd) | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETATE ION, CALCIUM ION, ... | | Authors: | Coban, M, Raeeszadeh-Sarmazdeh, M, Hockla, A, Sankaran, B, Radisky, E.S. | | Deposit date: | 2021-09-16 | | Release date: | 2022-03-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Engineering of tissue inhibitor of metalloproteinases TIMP-1 for fine discrimination between closely related stromelysins MMP-3 and MMP-10.
J.Biol.Chem., 298, 2022
|
|
7S7M
 
 | | Complex of tissue inhibitor of metalloproteinases-1 (TIMP-1) mutant (L34G/M66D/T98G/P131S/Q153N) with matrix metalloproteinase-3 catalytic domain (MMP-3cd) | | Descriptor: | CALCIUM ION, Metalloproteinase inhibitor 1, Stromelysin-1, ... | | Authors: | Coban, M, Raeeszadeh-Sarmazdeh, M, Sankaran, B, Hockla, A, Radisky, E.S. | | Deposit date: | 2021-09-16 | | Release date: | 2022-03-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Engineering of tissue inhibitor of metalloproteinases TIMP-1 for fine discrimination between closely related stromelysins MMP-3 and MMP-10.
J.Biol.Chem., 298, 2022
|
|
7Q0Y
 
 | | Crystal structure of CTX-M-14 in complex with Bortezomib | | Descriptor: | ACETATE ION, Beta-lactamase, CHLORIDE ION, ... | | Authors: | Werner, N, Perbandt, M, Hinrichs, W, Prester, A, Rohde, H, Aepfelbacher, M, Betzel, C. | | Deposit date: | 2021-10-17 | | Release date: | 2022-04-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structural basis to repurpose boron-based proteasome inhibitors Bortezomib and Ixazomib as beta-lactamase inhibitors.
Sci Rep, 12, 2022
|
|
7Q0Z
 
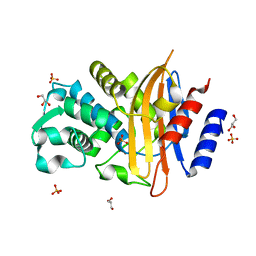 | | Crystal structure of CTX-M-14 | | Descriptor: | Beta-lactamase, GLYCEROL, SULFATE ION | | Authors: | Werner, N, Perbandt, M, Hinrichs, W, Prester, A, Rohde, H, Aepfelbacher, M, Betzel, C. | | Deposit date: | 2021-10-17 | | Release date: | 2022-04-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Structural basis to repurpose boron-based proteasome inhibitors Bortezomib and Ixazomib as beta-lactamase inhibitors.
Sci Rep, 12, 2022
|
|
7Q11
 
 | | Crystal structure of CTX-M-14 in complex with Ixazomib | | Descriptor: | Beta-lactamase, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Werner, N, Perbandt, M, Hinrichs, W, Prester, A, Rohde, H, Aepfelbacher, M, Betzel, C. | | Deposit date: | 2021-10-17 | | Release date: | 2022-04-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.14 Å) | | Cite: | Structural basis to repurpose boron-based proteasome inhibitors Bortezomib and Ixazomib as beta-lactamase inhibitors.
Sci Rep, 12, 2022
|
|
3R3L
 
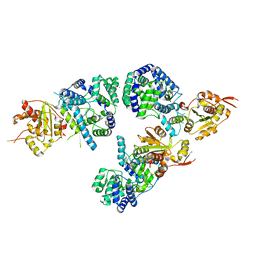 | | Structure of NP protein from Lassa AV strain | | Descriptor: | MANGANESE (II) ION, Nucleoprotein, ZINC ION | | Authors: | Perbandt, M, Brunotte, L, Gunther, S, Betzel, C. | | Deposit date: | 2011-03-16 | | Release date: | 2011-09-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.449 Å) | | Cite: | Structure of the Lassa virus nucleoprotein revealed by X-ray crystallography, small-angle X-ray scattering, and electron microscopy.
J.Biol.Chem., 286, 2011
|
|
6BX8
 
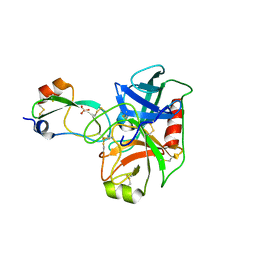 | | Human Mesotrypsin (PRSS3) Complexed with Tissue Factor Pathway Inhibitor Variant (TFPI1-KD1-K15R-I17C-I34C) | | Descriptor: | SULFATE ION, Tissue factor pathway inhibitor, Trypsin-3 | | Authors: | Coban, M, Sankaran, B, Cohen, I, Hockla, A, Papo, N, Radisky, E.S. | | Deposit date: | 2017-12-18 | | Release date: | 2019-02-06 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Disulfide engineering of human Kunitz-type serine protease inhibitors enhances proteolytic stability and target affinity toward mesotrypsin.
J. Biol. Chem., 294, 2019
|
|
6ITF
 
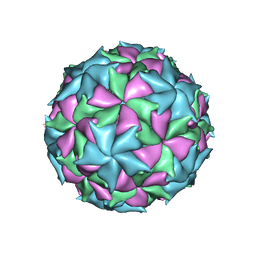 | |
6ITB
 
 | |
1QL0
 
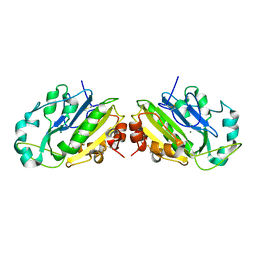 | |
3LOB
 
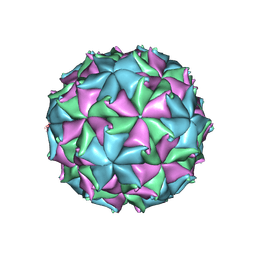 | | Crystal Structure of Flock House Virus calcium mutant | | Descriptor: | Coat protein beta, Coat protein gamma, RNA (5'-R(*UP*UP*U*AP*UP*CP*UP*(P))-3'), ... | | Authors: | Johnson, J.E, Banerjee, M, Speir, J.A, Huang, R. | | Deposit date: | 2010-02-03 | | Release date: | 2010-04-21 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Structure and function of a genetically engineered mimic of a nonenveloped virus entry intermediate.
J.Virol., 84, 2010
|
|
4E1Y
 
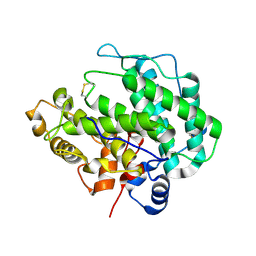 | | Alginate lyase A1-III H192A apo form | | Descriptor: | Alginate lyase | | Authors: | Mikami, B, Ban, M, Suzuki, S, Yoon, H.-J, Miyake, O, Yamasaki, M, Ogura, K, Maruyama, Y, Hashimoto, W, Murata, K. | | Deposit date: | 2012-03-07 | | Release date: | 2012-04-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Induced-fit motion of a lid loop involved in catalysis in alginate lyase A1-III
Acta Crystallogr.,Sect.D, 68, 2012
|
|
4F13
 
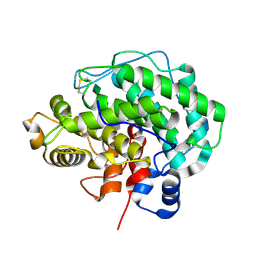 | | Alginate lyase A1-III Y246F complexed with tetrasaccharide | | Descriptor: | 4-deoxy-alpha-L-erythro-hex-4-enopyranuronic acid-(1-4)-beta-D-mannopyranuronic acid-(1-4)-beta-D-mannopyranuronic acid-(1-4)-beta-D-mannopyranuronic acid, Alginate lyase, beta-D-mannopyranuronic acid-(1-4)-beta-D-mannopyranuronic acid | | Authors: | Mikami, B, Ban, M, Suzuki, S, Yoon, H.-J, Miyake, O, Yamasaki, M, Ogura, K, Maruyama, Y, Hashimoto, W, Murata, K. | | Deposit date: | 2012-05-06 | | Release date: | 2012-06-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.208 Å) | | Cite: | Induced-fit motion of a lid loop involved in catalysis in alginate lyase A1-III
Acta Crystallogr.,Sect.D, 68, 2012
|
|
4F10
 
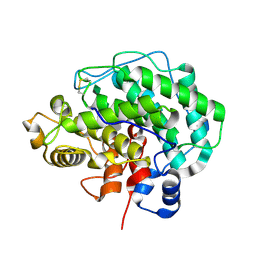 | | Alginate lyase A1-III H192A complexed with tetrasaccharide | | Descriptor: | 4-deoxy-alpha-L-erythro-hex-4-enopyranuronic acid-(1-4)-alpha-L-gulopyranuronic acid-(1-4)-beta-D-mannopyranuronic acid-(1-4)-beta-D-mannopyranuronic acid, Alginate lyase, alpha-L-gulopyranuronic acid-(1-4)-beta-D-mannopyranuronic acid | | Authors: | Mikami, B, Ban, M, Suzuki, S, Yoon, H.-J, Miyake, O, Yamasaki, M, Ogura, K, Maruyama, Y, Hashimoto, W, Murata, K. | | Deposit date: | 2012-05-05 | | Release date: | 2012-06-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Induced-fit motion of a lid loop involved in catalysis in alginate lyase A1-III
Acta Crystallogr.,Sect.D, 68, 2012
|
|
1JGC
 
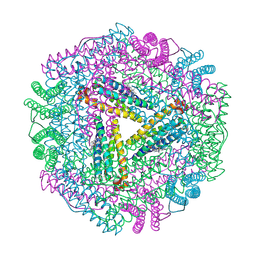 | | The 2.6 A Structure Resolution of Rhodobacter capsulatus Bacterioferritin with Metal-free Dinuclear Site and Heme Iron in a Crystallographic Special Position | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, bacterioferritin | | Authors: | Cobessi, D, Huang, L.-S, Ban, M, Pon, N.G, Daldal, F, Berry, E.A. | | Deposit date: | 2001-06-24 | | Release date: | 2002-01-09 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The 2.6 A resolution structure of Rhodobacter capsulatus bacterioferritin with metal-free dinuclear site and heme iron in a crystallographic 'special position'.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
4OW2
 
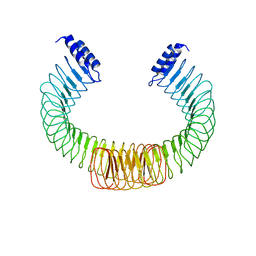 | | YopM from Yersinia enterocolitica WA-314 | | Descriptor: | Yop effector YopM | | Authors: | Rumm, A, Perbandt, M, Aepfelbacher, M. | | Deposit date: | 2014-01-30 | | Release date: | 2015-03-04 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Immunosuppressive Yersinia Effector YopM Binds DEAD Box Helicase DDX3 to Control Ribosomal S6 Kinase in the Nucleus of Host Cells.
PLoS Pathog., 12, 2016
|
|
3H9D
 
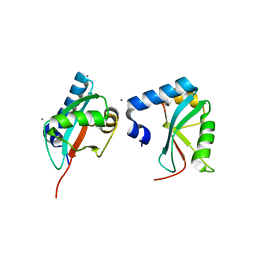 | | Crystal Structure of Trypanosoma brucei ATG8 | | Descriptor: | CALCIUM ION, Microtubule-associated protein 1A/1B, light chain 3, ... | | Authors: | Koopmann, R, Muhammad, K, Perbandt, M, Betzel, C, Duszenko, M. | | Deposit date: | 2009-04-30 | | Release date: | 2009-10-06 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Trypanosoma brucei ATG8: structural insights into autophagic-like mechanisms in protozoa.
Autophagy, 5, 2009
|
|
4UQF
 
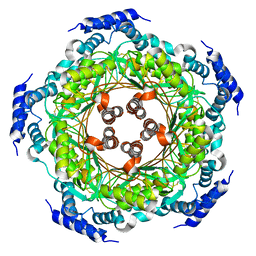 | | CRYSTAL STRUCTURE OF LISTERIA MONOCYTOGENES GTP CYCLOHYDROLASE I | | Descriptor: | GTP cyclohydrolase 1 | | Authors: | Schuessler, S, Perbandt, M, Fischer, M, Graewert, T. | | Deposit date: | 2014-06-23 | | Release date: | 2015-07-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of GTP cyclohydrolase I from Listeria monocytogenes, a potential anti-infective drug target.
Acta Crystallogr.,Sect.F, 75, 2019
|
|
1M9S
 
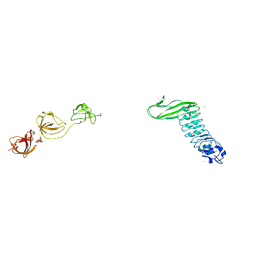 | | Crystal structure of Internalin B (InlB), a Listeria monocytogenes virulence protein containing SH3-like domains. | | Descriptor: | Internalin B, SULFATE ION, TERBIUM(III) ION | | Authors: | Marino, M, Banerjee, M, Jonquieres, R, Cossart, P, Ghosh, P. | | Deposit date: | 2002-07-29 | | Release date: | 2002-11-06 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | GW domains of the Listeria monocytogenes invasion protein InlB are SH3-like and mediate binding to host ligands
Embo J., 21, 2002
|
|
5D73
 
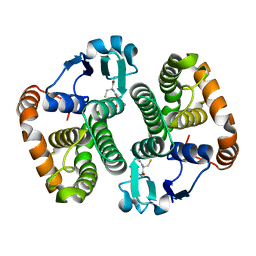 | | Structure of Wuchereria bancrofti pi-class glutathione S-transferase | | Descriptor: | GLUTATHIONE, Glutathione S-transferase | | Authors: | Prince, P.R, Sakthidevi, M, Madhumathi, J, Perbandt, M, Betzel, C, Kaliraj, P. | | Deposit date: | 2015-08-13 | | Release date: | 2015-11-11 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | STRUCTURE OF WUCHERERIA BANCROFTI PI-CLASS GLUTATHIONE S-TRANSFERASE
TO BE PUBLISHED
|
|
