2NOL
 
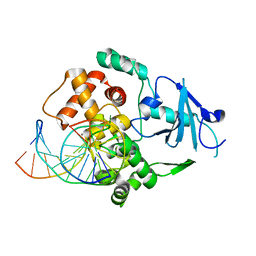 | | Structure of catalytically inactive human 8-oxoguanine glycosylase distal crosslink to oxoG DNA | | Descriptor: | 5'-D(*GP*CP*GP*TP*CP*CP*AP*(G42)P*GP*TP*CP*TP*AP*CP*C)-3', 5'-D(*GP*GP*TP*AP*GP*AP*CP*CP*TP*GP*GP*AP*CP*GP*C)-3', CALCIUM ION, ... | | Authors: | Banerjee, A, Radom, C.T, Verdine, G.L. | | Deposit date: | 2006-10-25 | | Release date: | 2006-11-21 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Structural characterization of human 8-oxoguanine DNA glycosylase variants bearing active site mutations.
J.Biol.Chem., 282, 2007
|
|
3GPU
 
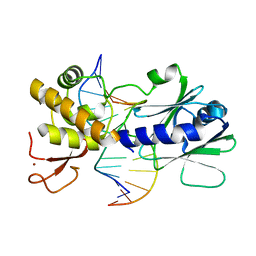 | | MutM encountering an intrahelical 8-oxoguanine (oxoG) lesion in EC4-loop deletion complex | | Descriptor: | DNA (5'-D(*AP*GP*GP*TP*AP*GP*AP*CP*TP*CP*GP*GP*AP*CP*GP*C)-3'), DNA (5'-D(*TP*GP*CP*GP*TP*CP*CP*(8OG)P*AP*GP*TP*CP*TP*AP*CP*C)-3'), DNA glycosylase, ... | | Authors: | Banerjee, A, Qi, Y, Verdine, G.L. | | Deposit date: | 2009-03-23 | | Release date: | 2009-11-10 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Encounter and extrusion of an intrahelical lesion by a DNA repair enzyme.
Nature, 462, 2009
|
|
3GQ3
 
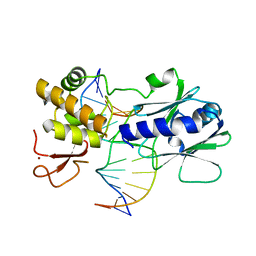 | | MutM encountering an intrahelical 8-oxoguanine (oxoG) lesion in EC5-loop deletion complex | | Descriptor: | DNA (5'-D(*AP*GP*GP*TP*AP*GP*AP*CP*CP*CP*GP*GP*AP*CP*GP*C)-3'), DNA (5'-D(*TP*GP*CP*GP*TP*CP*CP*GP*(8OG)P*GP*TP*CP*TP*AP*CP*C)-3'), DNA glycosylase, ... | | Authors: | Banerjee, A, Qi, Y, Verdine, G.L. | | Deposit date: | 2009-03-23 | | Release date: | 2009-11-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Encounter and extrusion of an intrahelical lesion by a DNA repair enzyme.
Nature, 462, 2009
|
|
1BQB
 
 | | AUREOLYSIN, STAPHYLOCOCCUS AUREUS METALLOPROTEINASE | | Descriptor: | CALCIUM ION, PROTEIN (AUREOLYSIN), ZINC ION | | Authors: | Medrano, F.J, Banbula, A, Potempa, J, Travis, J, Bode, W. | | Deposit date: | 1998-07-14 | | Release date: | 1999-01-13 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Amino-acid sequence and three-dimensional structure of the Staphylococcus aureus metalloproteinase at 1.72 A resolution.
Structure, 6, 1998
|
|
7ASD
 
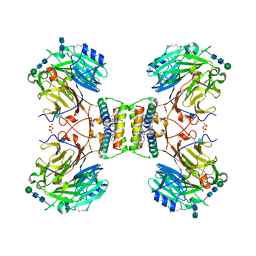 | | Structure of native royal jelly filaments | | Descriptor: | (3beta,14beta,17alpha)-ergosta-5,24(28)-dien-3-ol, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Mattei, S, Ban, A, Picenoni, A, Leibundgut, M, Glockshuber, R, Boehringer, D. | | Deposit date: | 2020-10-27 | | Release date: | 2020-12-30 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structure of native glycolipoprotein filaments in honeybee royal jelly.
Nat Commun, 11, 2020
|
|
5W7I
 
 | | X-ray structure of ankyrin repeat domain of DHHC17 in complex with Snap25b peptide | | Descriptor: | Palmitoyltransferase ZDHHC17, Snap25b-111-120 | | Authors: | Verardi, R, Kim, J.-S, Ghirlando, R, Banerjee, A. | | Deposit date: | 2017-06-20 | | Release date: | 2017-08-09 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.105 Å) | | Cite: | Structural Basis for Substrate Recognition by the Ankyrin Repeat Domain of Human DHHC17 Palmitoyltransferase.
Structure, 25, 2017
|
|
5WMB
 
 | | Structure of the 10S (-)-cis-BP-dG modified Rev1 ternary complex (the BP residue is disordered) | | Descriptor: | 1,2-ETHANEDIOL, 2'-DEOXYCYTIDINE-5'-TRIPHOSPHATE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Rechkoblit, O, Kolbanovsky, A, Landes, H, Geacintov, N.E, Aggarwal, A.K. | | Deposit date: | 2017-07-28 | | Release date: | 2017-10-25 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Mechanism of error-free replication across benzo[a]pyrene stereoisomers by Rev1 DNA polymerase.
Nat Commun, 8, 2017
|
|
5W7J
 
 | | X-ray structure of the E89A variant of ankyrin repeat domain of DHHC17 in complex with Snap25b peptide | | Descriptor: | Palmitoyltransferase ZDHHC17, Snap25b-111-120 | | Authors: | Verardi, R, Kim, J.-S, Ghirlando, R, Banerjee, A. | | Deposit date: | 2017-06-20 | | Release date: | 2017-08-09 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.202 Å) | | Cite: | Structural Basis for Substrate Recognition by the Ankyrin Repeat Domain of Human DHHC17 Palmitoyltransferase.
Structure, 25, 2017
|
|
2MIW
 
 | | Nuclear magnetic resonance studies of N2-guanine adducts derived from the tumorigen dibenzo[a,l]pyrene in DNA: Impact of adduct stereochemistry, size, and local DNA structure on solution conformations | | Descriptor: | (11R,12R,13R)-11,12,13,14-tetrahydronaphtho[1,2,3,4-pqr]tetraphene-11,12,13-triol, DNA_(5'-D(*CP*CP*AP*TP*CP*GP*CP*TP*AP*CP*C)-3'), DNA_(5'-D(*GP*GP*TP*AP*GP*CP*GP*AP*TP*GP*G)-3') | | Authors: | Rodriguez, F.A, Liu, Z, Lin, C.H, Ding, S, Cai, Y, Kolbanovskiy, A, Kolbanovskiy, M, Amin, S, Broyde, S, Geacintov, N.E. | | Deposit date: | 2013-12-20 | | Release date: | 2014-04-02 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Nuclear Magnetic Resonance Studies of an N(2)-Guanine Adduct Derived from the Tumorigen Dibenzo[a,l]pyrene in DNA: Impact of Adduct Stereochemistry, Size, and Local DNA Sequence on Solution Conformations.
Biochemistry, 53, 2014
|
|
2MIV
 
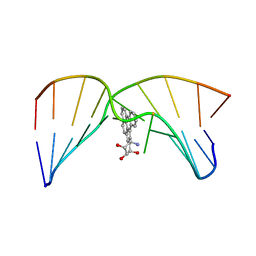 | | NMR studies of N2-guanine adducts derived from the tumorigen dibenzo[a,l]pyrene in DNA: Impact of adduct stereochemistry, size, and local DNA structure on solution conformations | | Descriptor: | (11R,12R,13R)-11,12,13,14-tetrahydronaphtho[1,2,3,4-pqr]tetraphene-11,12,13-triol, DNA_(5'-D(*CP*CP*AP*TP*CP*GP*CP*TP*AP*CP*C)-3'), DNA_(5'-D(*GP*GP*TP*AP*GP*GP*AP*TP*GP*G)-3') | | Authors: | Rodriguez, F.A, Liu, Z, Lin, C.H, Ding, S, Cai, Y, Kolbanovskiy, A, Kolbanovskiy, M, Amin, S, Broyde, S, Geacintov, N.E. | | Deposit date: | 2013-12-20 | | Release date: | 2014-04-02 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Nuclear Magnetic Resonance Studies of an N(2)-Guanine Adduct Derived from the Tumorigen Dibenzo[a,l]pyrene in DNA: Impact of Adduct Stereochemistry, Size, and Local DNA Sequence on Solution Conformations.
Biochemistry, 53, 2014
|
|
6WQZ
 
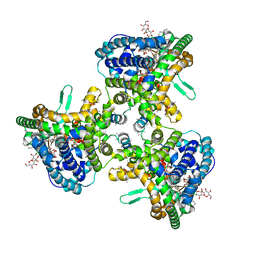 | | Structure of human ATG9A, the only transmembrane protein of the core autophagy machinery | | Descriptor: | Autophagy-related protein 9A, Lauryl Maltose Neopentyl Glycol | | Authors: | Guardia, C.M, Tan, X, Lian, T, Rana, M.S, Zhou, W, Christenson, E.T, Lowry, A.J, Faraldo-Gomez, J.D, Bonifacino, J.S, Jiang, J, Banerjee, A. | | Deposit date: | 2020-04-29 | | Release date: | 2020-07-08 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structure of Human ATG9A, the Only Transmembrane Protein of the Core Autophagy Machinery.
Cell Rep, 31, 2020
|
|
6WR4
 
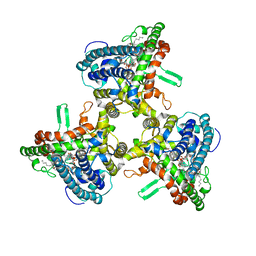 | | Structure of human ATG9A, the only transmembrane protein of the core autophagy machinery | | Descriptor: | Autophagy-related protein 9A, Lauryl Maltose Neopentyl Glycol | | Authors: | Guardia, C.M, Tan, X, Lian, T, Rana, M.S, Zhou, W, Christenson, E.T, Lowry, A.J, Faraldo-Gomez, J.D, Bonifacino, J.S, Jiang, J, Banerjee, A. | | Deposit date: | 2020-04-29 | | Release date: | 2020-07-08 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structure of Human ATG9A, the Only Transmembrane Protein of the Core Autophagy Machinery.
Cell Rep, 31, 2020
|
|
8XZ3
 
 | |
6I9A
 
 | | Porphyromonas gingivalis gingipain K (Kgp) in complex with inhibitor KYT-36 | | Descriptor: | (2S,3S)-1,4-DIMERCAPTOBUTANE-2,3-DIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Gomis-Ruth, F.X, Guevara, T, Rofdriguez-Banqueri, A. | | Deposit date: | 2018-11-22 | | Release date: | 2019-03-13 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structural determinants of inhibition of Porphyromonas gingivalis gingipain K by KYT-36, a potent, selective, and bioavailable peptidase inhibitor.
Sci Rep, 9, 2019
|
|
5ZHP
 
 | | M3 muscarinic acetylcholine receptor in complex with a selective antagonist | | Descriptor: | (1R,2R,4S,5S,7s)-7-({[4-fluoro-2-(thiophen-2-yl)phenyl]carbamoyl}oxy)-9,9-dimethyl-3-oxa-9-azatricyclo[3.3.1.0~2,4~]nonan-9-ium, CITRIC ACID, HEXAETHYLENE GLYCOL, ... | | Authors: | Liu, H, Hofmann, J, Fish, I, Schaake, B, Eitel, K, Bartuschat, A, Kaindl, J, Rampp, H, Banerjee, A, Hubner, H, Clark, M.J, Vincent, S.G, Fisher, J, Heinrich, M, Hirata, K, Liu, X, Sunahara, R.K, Shoichet, B.K, Kobilka, B.K, Gmeiner, P. | | Deposit date: | 2018-03-13 | | Release date: | 2018-11-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure-guided development of selective M3 muscarinic acetylcholine receptor antagonists
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
7NEN
 
 | |
7SKL
 
 | | Complex between S. aureus aureolysin and IMPI mutant I57I | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, IMPI alpha, ... | | Authors: | Mendes, S.R, Eckhard, U, Rodriguez-Banqueri, A, Guevara, T, Gomis-Ruth, F.X. | | Deposit date: | 2021-10-21 | | Release date: | 2022-01-26 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | An engineered protein-based submicromolar competitive inhibitor of the Staphylococcus aureus virulence factor aureolysin
Comput Struct Biotechnol J, 20, 2022
|
|
7SKM
 
 | | Complex between S. aureus aureolysin and wt IMPI. | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Mendes, S.R, Eckhard, U, Rodriguez-Banqueri, A, Guevara, T, Gomis-Ruth, F.X. | | Deposit date: | 2021-10-21 | | Release date: | 2022-01-26 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | An engineered protein-based submicromolar competitive inhibitor of the Staphylococcus aureus virulence factor aureolysin
Comput Struct Biotechnol J, 20, 2022
|
|
5C1V
 
 | | CRYSTAL STRUCTURE ANALYSIS OF CATALYTIC SUBUNIT OF HUMAN CALCINEURIN | | Descriptor: | FE (III) ION, PHOSPHATE ION, Serine/threonine-protein phosphatase 2B catalytic subunit alpha isoform, ... | | Authors: | Guasch, A, Fita, I, Perez-Luque, R, Aparicio, D, Aranguren-Ibanez, A, Perez-Riba, M. | | Deposit date: | 2015-06-15 | | Release date: | 2016-02-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | Calcineurin Undergoes a Conformational Switch Evoked via Peptidyl-Prolyl Isomerization.
Plos One, 10, 2015
|
|
7BML
 
 | |
8CD8
 
 | | Ulilysin - C269A with AEBSF complex | | Descriptor: | 4-(2-AMINOETHYL)BENZENESULFONYL FLUORIDE, CALCIUM ION, GLY-SER-SER, ... | | Authors: | Rodriguez-Banqueri, A, Eckhard, U, Gomis-Ruth, F.X. | | Deposit date: | 2023-01-30 | | Release date: | 2023-03-22 | | Last modified: | 2023-03-29 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural insights into latency of the metallopeptidase ulilysin (lysargiNase) and its unexpected inhibition by a sulfonyl-fluoride inhibitor of serine peptidases.
Dalton Trans, 52, 2023
|
|
7Z5E
 
 | | VP2-only capsid of MVM D263A mutant | | Descriptor: | Capsid protein VP1 | | Authors: | Luque, D, Ortega-Esteban, A, Valbuena, A, Vilas, J.L, Rodriguez-Huete, A, Mateu, M.G, Caston, J.R. | | Deposit date: | 2022-03-09 | | Release date: | 2023-03-08 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.32 Å) | | Cite: | Equilibrium Dynamics of a Biomolecular Complex Analyzed at Single-amino Acid Resolution by Cryo-electron Microscopy.
J.Mol.Biol., 435, 2023
|
|
7Z5F
 
 | | VP2-only capsid of MVM D263A mutant | | Descriptor: | Capsid protein VP1 | | Authors: | Luque, D, Ortega-Esteban, A, Valbuena, A, Vilas, J.L, Rodriguez-Huete, A, Mateu, M.G, Caston, J.R. | | Deposit date: | 2022-03-09 | | Release date: | 2023-03-08 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.22 Å) | | Cite: | Equilibrium Dynamics of a Biomolecular Complex Analyzed at Single-amino Acid Resolution by Cryo-electron Microscopy.
J.Mol.Biol., 435, 2023
|
|
7Z5D
 
 | | VP2-only capsid of wt MVM prototype strain p | | Descriptor: | Capsid protein VP1 | | Authors: | Luque, D, Ortega-Esteban, A, Valbuena, A, Vilas, J.L, Rodriguez-Huete, A, Mateu, M.G, Caston, J.R. | | Deposit date: | 2022-03-09 | | Release date: | 2023-03-08 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.42 Å) | | Cite: | Equilibrium Dynamics of a Biomolecular Complex Analyzed at Single-amino Acid Resolution by Cryo-electron Microscopy.
J.Mol.Biol., 435, 2023
|
|
8P8N
 
 | | Mouse RPL39 integrated into the yeast 60S ribosomal subunit | | Descriptor: | 25S rRNA, 5.8S rRNA, 5S rRNA, ... | | Authors: | Rabl, J, Banerjee, A, Boehringer, D, Zavolan, M. | | Deposit date: | 2023-06-02 | | Release date: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.15 Å) | | Cite: | Yeast 60S ribosomal subunit, RPL39 deletion
To Be Published
|
|
