6HXL
 
 | |
6HXH
 
 | | Structure of the human ATP citrate lyase holoenzyme in complex with citrate, coenzyme A and Mg.ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-citrate synthase,Human ATP citrate lyase, CITRATE ANION, ... | | Authors: | Verstraete, K, Verschueren, K. | | Deposit date: | 2018-10-17 | | Release date: | 2019-04-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structure of ATP citrate lyase and the origin of citrate synthase in the Krebs cycle.
Nature, 568, 2019
|
|
6HXN
 
 | |
6HXP
 
 | |
1I2S
 
 | | BETA-LACTAMASE FROM BACILLUS LICHENIFORMIS BS3 | | Descriptor: | BETA-LACTAMASE, CITRIC ACID, SODIUM ION | | Authors: | Fonze, E, Vanhove, M, Dive, G, Sauvage, E, Frere, J.M, Charlier, P. | | Deposit date: | 2001-02-12 | | Release date: | 2002-03-13 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structures of the Bacillus licheniformis BS3 class A beta-lactamase and of the
acyl-enzyme adduct formed with cefoxitin
Biochemistry, 41, 2002
|
|
1I2W
 
 | | BETA-LACTAMASE FROM BACILLUS LICHENIFORMIS BS3 COMPLEXED WITH CEFOXITIN | | Descriptor: | (2R)-5-[(carbamoyloxy)methyl]-2-{(1S)-1-methoxy-2-oxo-1-[(thiophen-2-ylacetyl)amino]ethyl}-3,6-dihydro-2H-1,3-thiazine-4-carboxylic acid, BETA-LACTAMASE, CARBAMIC ACID | | Authors: | Fonze, E, Vanhove, M, Dive, G, Sauvage, E, Frere, J.M, Charlier, P. | | Deposit date: | 2001-02-12 | | Release date: | 2002-03-13 | | Last modified: | 2018-09-12 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structures of the Bacillus licheniformis BS3 class A beta-lactamase and of the acyl-enzyme adduct formed with cefoxitin
Biochemistry, 41, 2002
|
|
2GOU
 
 | | Structure of wild type, oxidized SYE1, an OYE homologue from S. oneidensis | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Savvides, S.N, van den Hemel, D. | | Deposit date: | 2006-04-14 | | Release date: | 2006-07-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Ligand-induced conformational changes in the capping subdomain of a bacterial old yellow enzyme homologue and conserved sequence fingerprints provide new insights into substrate binding.
J.Biol.Chem., 281, 2006
|
|
2GQA
 
 | | Structure of NADH-reduced SYE1, an OYE homologue from S. oneidensis | | Descriptor: | FLAVIN MONONUCLEOTIDE, SULFATE ION, oxidoreductase, ... | | Authors: | Savvides, S.N, van den Hemel, D. | | Deposit date: | 2006-04-20 | | Release date: | 2006-07-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Ligand-induced conformational changes in the capping subdomain of a bacterial old yellow enzyme homologue and conserved sequence fingerprints provide new insights into substrate binding.
J.Biol.Chem., 281, 2006
|
|
1G9K
 
 | |
2X0N
 
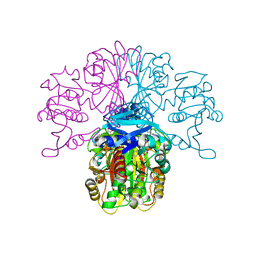 | | Structure of glycosomal glyceraldehyde-3-phosphate dehydrogenase from Trypanosoma brucei determined from Laue data | | Descriptor: | GLYCERALDEHYDE-3-PHOSPHATE DEHYDROGENASE, GLYCOSOMAL, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Vellieux, F.M.D, Hajdu, J, Hol, W.G.J. | | Deposit date: | 2009-12-16 | | Release date: | 2009-12-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structure of Glycosomal Glyceraldehyde-3-Phosphate Dehydrogenase from Trypanosoma Brucei Determined from Laue Data.
Proc.Natl.Acad.Sci.USA, 90, 1993
|
|
2GQ9
 
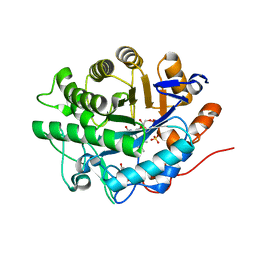 | | Structure of SYE1, an OYE homologue from S. oneidensis, in complex with p-hydroxybenzaldehyde | | Descriptor: | FLAVIN MONONUCLEOTIDE, P-HYDROXYBENZALDEHYDE, SULFATE ION, ... | | Authors: | Savvides, S.N, van den Hemel, D. | | Deposit date: | 2006-04-20 | | Release date: | 2006-07-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Ligand-induced conformational changes in the capping subdomain of a bacterial old yellow enzyme homologue and conserved sequence fingerprints provide new insights into substrate binding.
J.Biol.Chem., 281, 2006
|
|
2GQ8
 
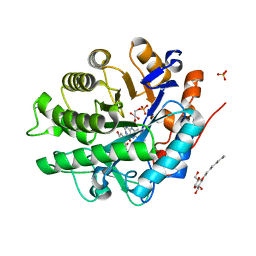 | | Structure of SYE1, an OYE homologue from S. ondeidensis, in complex with p-hydroxyacetophenone | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, FLAVIN MONONUCLEOTIDE, P-HYDROXYACETOPHENONE, ... | | Authors: | Savvides, S.N, van den Hemel, D. | | Deposit date: | 2006-04-20 | | Release date: | 2006-07-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Ligand-induced conformational changes in the capping subdomain of a bacterial old yellow enzyme homologue and conserved sequence fingerprints provide new insights into substrate binding.
J.Biol.Chem., 281, 2006
|
|
1HIX
 
 | | CRYSTALLOGRAPHIC ANALYSES OF FAMILY 11 ENDO-BETA-1,4-XYLANASE XYL1 FROM STREPTOMYCES SP. S38 | | Descriptor: | ENDO-1,4-BETA-XYLANASE | | Authors: | Wouters, J, Georis, J, Dusart, J, Frere, J.M, Depiereux, E, Charlier, P. | | Deposit date: | 2001-01-05 | | Release date: | 2001-11-30 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystallographic Analysis of Family 11 Endo-[Beta]-1,4-Xylanase Xyl1 from Streptomyces Sp. S38
Acta Crystallogr.,Sect.D, 57, 2001
|
|
2QZ6
 
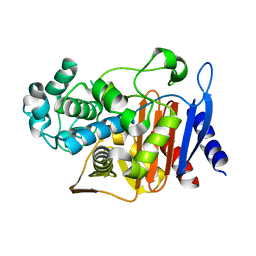 | | First crystal structure of a psychrophile class C beta-lactamase | | Descriptor: | Beta-lactamase | | Authors: | Michaux, C, Massant, J, Kerff, F, Charlier, P, Wouters, J. | | Deposit date: | 2007-08-16 | | Release date: | 2008-03-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Crystal structure of a cold-adapted class C beta-lactamase
Febs J., 275, 2008
|
|
1J9M
 
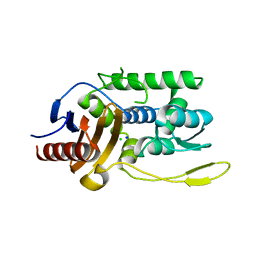 | | K38H mutant of Streptomyces K15 DD-transpeptidase | | Descriptor: | CHLORIDE ION, DD-transpeptidase, SODIUM ION | | Authors: | Fonze, E, Rhazi, N, Nguyen-Disteche, M, Charlier, P. | | Deposit date: | 2001-05-28 | | Release date: | 2001-06-13 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Catalytic mechanism of the Streptomyces K15 DD-transpeptidase/penicillin-binding protein probed by site-directed mutagenesis and structural analysis.
Biochemistry, 42, 2003
|
|
3PHY
 
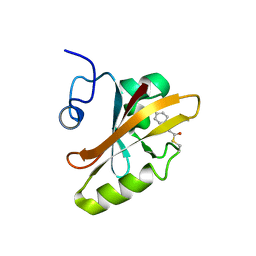 | | PHOTOACTIVE YELLOW PROTEIN, DARK STATE (UNBLEACHED), SOLUTION STRUCTURE, NMR, 26 STRUCTURES | | Descriptor: | 4'-HYDROXYCINNAMIC ACID, PHOTOACTIVE YELLOW PROTEIN | | Authors: | Dux, P, Rubinstenn, G, Vuister, G.W, Boelens, R, Mulder, F.A.A, Hard, K, Hoff, W.D, Kroon, A, Crielaard, W, Hellingwerf, K.J, Kaptein, R. | | Deposit date: | 1998-02-06 | | Release date: | 1998-05-27 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | Solution structure and backbone dynamics of the photoactive yellow protein.
Biochemistry, 37, 1998
|
|
1W79
 
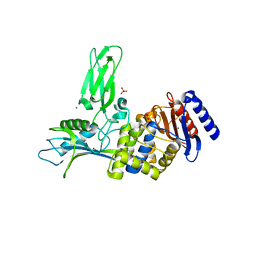 | | Crystal structure of the DD-transpeptidase-carboxypeptidase from Actinomadura R39 | | Descriptor: | D-alanyl-D-alanine carboxypeptidase, MAGNESIUM ION, SULFATE ION | | Authors: | Sauvage, E, Herman, R, Petrella, S, Duez, C, Frere, J.M, Charlier, P. | | Deposit date: | 2004-08-31 | | Release date: | 2005-06-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of the Actinomadura R39 DD-peptidase reveals new domains in penicillin-binding proteins.
J. Biol. Chem., 280, 2005
|
|
3ERX
 
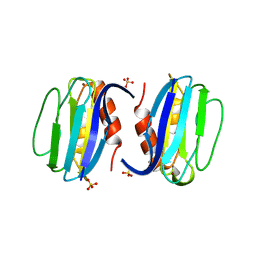 | | High-resolution structure of Paracoccus pantotrophus pseudoazurin | | Descriptor: | COPPER (II) ION, Pseudoazurin, SULFATE ION | | Authors: | Najmudin, S, Pauleta, S.R, Moura, I, Romao, M.J. | | Deposit date: | 2008-10-03 | | Release date: | 2009-10-13 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | The 1.4 A resolution structure of Paracoccus pantotrophus pseudoazurin.
Acta Crystallogr.,Sect.F, 66, 2010
|
|
1ES4
 
 | | C98N mutant of streptomyces K15 DD-transpeptidase | | Descriptor: | DD-TRANSPEPTIDASE | | Authors: | Fonze, E, Charlier, P. | | Deposit date: | 2000-04-07 | | Release date: | 2000-05-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Catalytic mechanism of the Streptomyces K15 DD-transpeptidase/penicillin-binding protein probed by site-directed mutagenesis and structural analysis.
Biochemistry, 42, 2003
|
|
1ES5
 
 | |
1ES2
 
 | | S96A mutant of streptomyces K15 DD-transpeptidase | | Descriptor: | DD-TRANSPEPTIDASE | | Authors: | Fonze, E, Charlier, P. | | Deposit date: | 2000-04-07 | | Release date: | 2000-05-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Catalytic mechanism of the Streptomyces K15 DD-transpeptidase/penicillin-binding protein probed by site-directed mutagenesis and structural analysis.
Biochemistry, 42, 2003
|
|
1ES3
 
 | | C98A mutant of streptomyces K15 DD-transpeptidase | | Descriptor: | DD-TRANSPEPTIDASE, SODIUM ION | | Authors: | Fonze, E, Charlier, P. | | Deposit date: | 2000-04-07 | | Release date: | 2000-05-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Catalytic mechanism of the Streptomyces K15 DD-transpeptidase/penicillin-binding protein probed by site-directed mutagenesis and structural analysis.
Biochemistry, 42, 2003
|
|
1ESI
 
 | |
1SKF
 
 | | CRYSTAL STRUCTURE OF THE STREPTOMYCES K15 DD-TRANSPEPTIDASE | | Descriptor: | D-ALANYL-D-ALANINE TRANSPEPTIDASE | | Authors: | Fonze, E, Charlier, P. | | Deposit date: | 1998-08-20 | | Release date: | 1999-08-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The crystal structure of a penicilloyl-serine transferase of intermediate penicillin sensitivity. The DD-transpeptidase of streptomyces K15.
J.Biol.Chem., 274, 1999
|
|
1GZJ
 
 | |
