8VTT
 
 | |
8VTS
 
 | |
6MYK
 
 | | Pleurotus ostreatus OstreolysinA mutant E69A with Bis-Tris | | Descriptor: | 1,2-ETHANEDIOL, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Ostreolysin A6, ... | | Authors: | Tomchick, D.R, Radhakrishnan, A, Endapally, S. | | Deposit date: | 2018-11-01 | | Release date: | 2019-02-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Molecular Discrimination between Two Conformations of Sphingomyelin in Plasma Membranes.
Cell, 176, 2019
|
|
6MYJ
 
 | | Pleurotus ostreatus OstreolysinA plus sphingomyelin | | Descriptor: | 1,2-ETHANEDIOL, N-[(2S)-1-hydroxypropan-2-yl]butanamide, Ostreolysin A6, ... | | Authors: | Tomchick, D.R, Radhakrishnan, A, Endapally, S. | | Deposit date: | 2018-11-01 | | Release date: | 2019-02-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | Molecular Discrimination between Two Conformations of Sphingomyelin in Plasma Membranes.
Cell, 176, 2019
|
|
6MYI
 
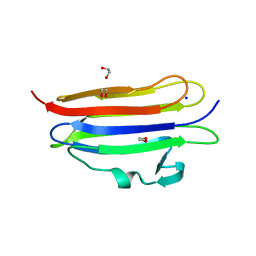 | | Pleurotus ostreatus OstreolysinA | | Descriptor: | 1,2-ETHANEDIOL, Ostreolysin A6, SODIUM ION | | Authors: | Tomchick, D.R, Radhakrishnan, A, Endapally, S. | | Deposit date: | 2018-11-01 | | Release date: | 2019-02-13 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Molecular Discrimination between Two Conformations of Sphingomyelin in Plasma Membranes.
Cell, 176, 2019
|
|
6N1Z
 
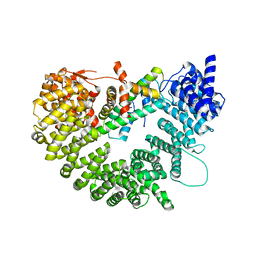 | | Importin-9 bound to H2A-H2B | | Descriptor: | Histone H2A, Histone H2B 1.1, Importin-9 | | Authors: | Tomchick, D.R, Chook, Y.M, Padavannil, A. | | Deposit date: | 2018-11-12 | | Release date: | 2019-03-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Importin-9 wraps around the H2A-H2B core to act as nuclear importer and histone chaperone.
Elife, 8, 2019
|
|
6NYT
 
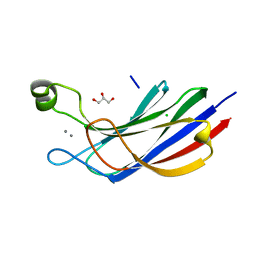 | | Munc13-1 C2B-domain, calcium bound | | Descriptor: | CALCIUM ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Tomchick, D.R, Rizo, J, Machius, M, Lu, J. | | Deposit date: | 2019-02-12 | | Release date: | 2019-02-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.369 Å) | | Cite: | Munc13 C2B domain is an activity-dependent Ca2+ regulator of synaptic exocytosis.
Nat. Struct. Mol. Biol., 17, 2010
|
|
6NYC
 
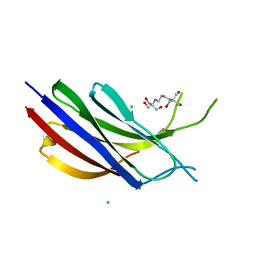 | | Munc13-1 C2B-domain, calcium free | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, Munc13-1 | | Authors: | Tomchick, D.R, Rizo, J, Machius, M, Lu, J. | | Deposit date: | 2019-02-11 | | Release date: | 2019-02-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.893 Å) | | Cite: | Munc13 C2B domain is an activity-dependent Ca2+ regulator of synaptic exocytosis.
Nat. Struct. Mol. Biol., 17, 2010
|
|
6OQQ
 
 | | Legionella pneumophila SidJ/Saccharomyces cerevisiae calmodulin complex | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE MONOPHOSPHATE, CALCIUM ION, ... | | Authors: | Tomchick, D.R, Tagliabracci, V.S, Black, M, Osinski, A. | | Deposit date: | 2019-04-28 | | Release date: | 2019-05-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.102 Å) | | Cite: | Bacterial pseudokinase catalyzes protein polyglutamylation to inhibit the SidE-family ubiquitin ligases.
Science, 364, 2019
|
|
5EW4
 
 | | Crystal structure of C9ORF72 Antisense CCCCGG repeat RNA associated with Lou Gehrig's disease and frontotemporal dementia, crystallized with Sr2+ | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, POTASSIUM ION, ... | | Authors: | Tomchick, D.R, Dodd, D.W, Gagnon, K.T, Corey, D.R. | | Deposit date: | 2015-11-20 | | Release date: | 2016-02-24 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Pathogenic C9ORF72 Antisense Repeat RNA Forms a Double Helix with Tandem C:C Mismatches.
Biochemistry, 55, 2016
|
|
5EW7
 
 | | Crystal structure of C9ORF72 Antisense CCCCGG repeat RNA associated with Lou Gehrig's disease and frontotemporal dementia, crystallized with Ba2+ | | Descriptor: | BARIUM ION, MAGNESIUM ION, POTASSIUM ION, ... | | Authors: | Tomchick, D.R, Dodd, D.W, Gagnon, K.T, Corey, D.R. | | Deposit date: | 2015-11-20 | | Release date: | 2016-02-24 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Pathogenic C9ORF72 Antisense Repeat RNA Forms a Double Helix with Tandem C:C Mismatches.
Biochemistry, 55, 2016
|
|
5UF7
 
 | | CRYSTAL STRUCTURE OF MUNC13-1 MUN DOMAIN | | Descriptor: | Protein unc-13 homolog A | | Authors: | Tomchick, D.R, Rizo, J, Xu, J. | | Deposit date: | 2017-01-03 | | Release date: | 2017-02-15 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.896 Å) | | Cite: | Mechanistic insights into neurotransmitter release and presynaptic plasticity from the crystal structure of Munc13-1 C1C2BMUN.
Elife, 6, 2017
|
|
5UE8
 
 | | The crystal structure of Munc13-1 C1C2BMUN domain | | Descriptor: | CHLORIDE ION, Protein unc-13 homolog A, ZINC ION | | Authors: | Tomchick, D.R, Rizo, J, Xu, J. | | Deposit date: | 2016-12-29 | | Release date: | 2017-02-15 | | Last modified: | 2018-03-28 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | Mechanistic insights into neurotransmitter release and presynaptic plasticity from the crystal structure of Munc13-1 C1C2BMUN.
Elife, 6, 2017
|
|
3L15
 
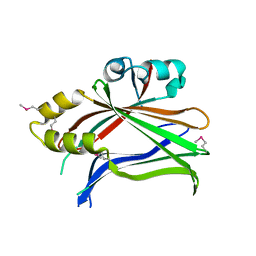 | | Human Tead2 transcriptional factor | | Descriptor: | GLYCEROL, Transcriptional enhancer factor TEF-4 | | Authors: | Tomchick, D.R, Luo, X, Tian, W. | | Deposit date: | 2009-12-10 | | Release date: | 2010-04-07 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and functional analysis of the YAP-binding domain of human TEAD2.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3KY9
 
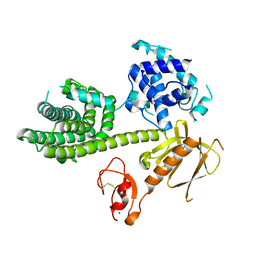 | | Autoinhibited Vav1 | | Descriptor: | Proto-oncogene vav, ZINC ION | | Authors: | Tomchick, D.R, Rosen, M.K, Machius, M, Yu, B. | | Deposit date: | 2009-12-04 | | Release date: | 2010-02-23 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.731 Å) | | Cite: | Structural and Energetic Mechanisms of Cooperative Autoinhibition and Activation of Vav1
Cell(Cambridge,Mass.), 140, 2010
|
|
3PCR
 
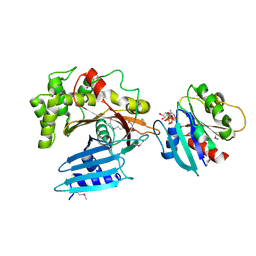 | | Structure of EspG-Arf6 complex | | Descriptor: | ADP-ribosylation factor 6, EspG, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Tomchick, D.R, Alto, N.M, Selyunin, A.S. | | Deposit date: | 2010-10-21 | | Release date: | 2011-01-05 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The assembly of a GTPase-kinase signalling complex by a bacterial catalytic scaffold.
Nature, 469, 2011
|
|
3N5A
 
 | | Synaptotagmin-7, C2B-domain, calcium bound | | Descriptor: | CALCIUM ION, Synaptotagmin-7 | | Authors: | Tomchick, D.R, Rizo, J, Craig, T.K. | | Deposit date: | 2010-05-24 | | Release date: | 2010-09-29 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.441 Å) | | Cite: | Structural and mutational analysis of functional differentiation between synaptotagmins-1 and -7.
Plos One, 5, 2010
|
|
3PCS
 
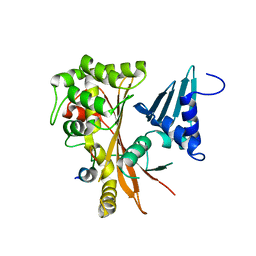 | |
3OBV
 
 | | Autoinhibited Formin mDia1 Structure | | Descriptor: | Protein diaphanous homolog 1, beta-D-fructofuranose-(2-1)-alpha-D-glucopyranose | | Authors: | Tomchick, D.R, Rosen, M.K, Otomo, T. | | Deposit date: | 2010-08-09 | | Release date: | 2010-11-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Crystal structure of the Formin mDia1 in autoinhibited conformation.
Plos One, 5, 2010
|
|
4NPK
 
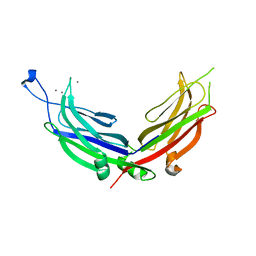 | | Extended-Synaptotagmin 2, C2A- and C2B-domains, calcium bound | | Descriptor: | CALCIUM ION, Extended synaptotagmin-2 | | Authors: | Tomchick, D.R, Rizo, J, Xu, J. | | Deposit date: | 2013-11-21 | | Release date: | 2014-01-29 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.552 Å) | | Cite: | Structure and ca(2+)-binding properties of the tandem c2 domains of e-syt2.
Structure, 22, 2014
|
|
4NPJ
 
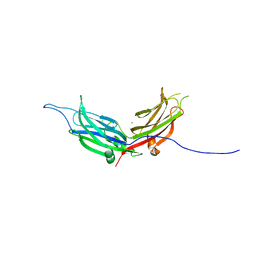 | | Extended-Synaptotagmin 2, C2A- and C2B-domains | | Descriptor: | ACETATE ION, CHLORIDE ION, Extended synaptotagmin-2, ... | | Authors: | Tomchick, D.R, Rizo, J, Xu, J. | | Deposit date: | 2013-11-21 | | Release date: | 2014-01-29 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.101 Å) | | Cite: | Structure and ca(2+)-binding properties of the tandem c2 domains of e-syt2.
Structure, 22, 2014
|
|
4PKY
 
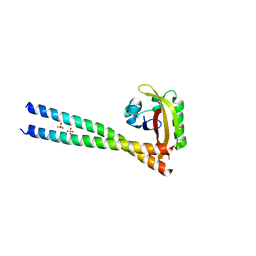 | | ARNT/HIF transcription factor/coactivator complex | | Descriptor: | Aryl hydrocarbon receptor nuclear translocator, Endothelial PAS domain-containing protein 1, SULFATE ION, ... | | Authors: | Tomchick, D.R, Partch, C.L, Gardner, K.H. | | Deposit date: | 2014-05-15 | | Release date: | 2015-02-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Coiled-coil Coactivators Play a Structural Role Mediating Interactions in Hypoxia-inducible Factor Heterodimerization.
J.Biol.Chem., 290, 2015
|
|
4ONR
 
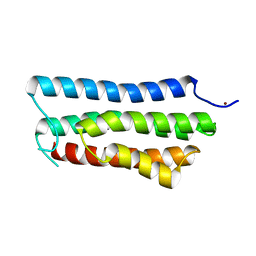 | |
4R39
 
 | |
4IFX
 
 | | Crystal structure of Treponema pallidum TP0796 Flavin trafficking protein, FAD substrate bound form | | Descriptor: | ACETATE ION, FLAVIN-ADENINE DINUCLEOTIDE, MAGNESIUM ION, ... | | Authors: | Tomchick, D.R, Brautigam, C.A, Deka, R.K, Norgard, M.V. | | Deposit date: | 2012-12-15 | | Release date: | 2013-02-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.452 Å) | | Cite: | The TP0796 Lipoprotein of Treponema pallidum Is a Bimetal-dependent FAD Pyrophosphatase with a Potential Role in Flavin Homeostasis.
J.Biol.Chem., 288, 2013
|
|
