3F1I
 
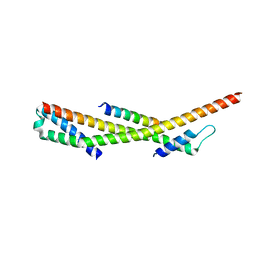 | | Human ESCRT-0 Core Complex | | Descriptor: | Hepatocyte growth factor-regulated tyrosine kinase substrate, Signal transducing adapter molecule 1 | | Authors: | Ren, X, Kloer, D.P, Kim, Y, Ghirlando, R, Saidi, L, Hummer, G, Hurley, J.H. | | Deposit date: | 2008-10-28 | | Release date: | 2009-03-24 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Hybrid Structural Model of the Complete Human ESCRT-0 Complex.
Structure, 17, 2009
|
|
6CM9
 
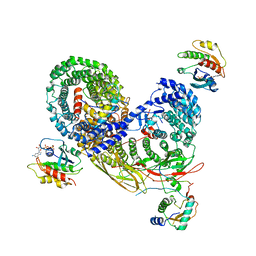 | | Structure of the cargo bound AP-1:Arf1:tetherin-Nef closed trimer monomeric subunit | | Descriptor: | ADP-ribosylation factor 1, AP-1 complex subunit beta-1, AP-1 complex subunit gamma-1, ... | | Authors: | Morris, K.L, Buffalo, C.Z, Ren, X, Hurley, J.H. | | Deposit date: | 2018-03-03 | | Release date: | 2018-08-08 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.73 Å) | | Cite: | HIV-1 Nefs Are Cargo-Sensitive AP-1 Trimerization Switches in Tetherin Downregulation.
Cell, 174, 2018
|
|
6CRI
 
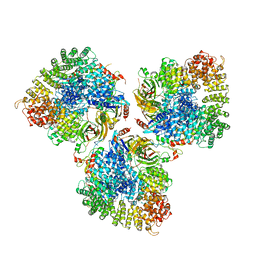 | | Structure of the cargo bound AP-1:Arf1:tetherin-Nef stable closed trimer | | Descriptor: | ADP-ribosylation factor 1, AP-1 complex subunit beta-1, AP-1 complex subunit gamma-1, ... | | Authors: | Morris, K.L, Buffalo, C.Z, Hurley, J.H. | | Deposit date: | 2018-03-18 | | Release date: | 2018-08-01 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (6.8 Å) | | Cite: | HIV-1 Nefs Are Cargo-Sensitive AP-1 Trimerization Switches in Tetherin Downregulation.
Cell, 174, 2018
|
|
3E1R
 
 | | Midbody targeting of the ESCRT machinery by a non-canonical coiled-coil in CEP55 | | Descriptor: | Centrosomal protein of 55 kDa, Programmed cell death 6-interacting protein | | Authors: | Lee, H.H, Elia, N, Ghirlando, R, Lippincott-Schwartz, J, Hurley, J.H. | | Deposit date: | 2008-08-04 | | Release date: | 2008-11-04 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Midbody targeting of the ESCRT machinery by a noncanonical coiled coil in CEP55.
Science, 322, 2008
|
|
6DCE
 
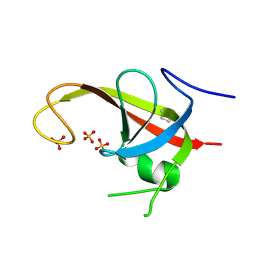 | | X-ray structure of FIP200 claw domain | | Descriptor: | RB1-inducible coiled-coil protein 1, SULFATE ION | | Authors: | Su, M.-Y, Hurley, J.H. | | Deposit date: | 2018-05-05 | | Release date: | 2019-03-06 | | Last modified: | 2019-05-01 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | FIP200 Claw Domain Binding to p62 Promotes Autophagosome Formation at Ubiquitin Condensates.
Mol. Cell, 74, 2019
|
|
6D84
 
 | | Structure of the cargo bound AP-1:Arf1:tetherin-Nef (L164A, L165A) dileucine mutant dimer | | Descriptor: | ADP-ribosylation factor 1, AP-1 complex subunit beta-1, AP-1 complex subunit gamma-1, ... | | Authors: | Buffalo, C.Z, Morris, K.L, Hurley, J.H. | | Deposit date: | 2018-04-25 | | Release date: | 2018-08-08 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (6.72 Å) | | Cite: | HIV-1 Nefs Are Cargo-Sensitive AP-1 Trimerization Switches in Tetherin Downregulation.
Cell, 174, 2018
|
|
6DFF
 
 | | Structure of the cargo bound AP-1:Arf1:tetherin-Nef monomer | | Descriptor: | ADP-ribosylation factor 1, AP-1 complex subunit beta-1, AP-1 complex subunit gamma-1, ... | | Authors: | Morris, K.L, Buffalo, C.Z, Ren, X, Hurley, J.H. | | Deposit date: | 2018-05-14 | | Release date: | 2018-08-08 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | HIV-1 Nefs Are Cargo-Sensitive AP-1 Trimerization Switches in Tetherin Downregulation.
Cell, 174, 2018
|
|
6NZD
 
 | | Cryo-EM Structure of the Lysosomal Folliculin Complex (FLCN-FNIP2-RagA-RagC-Ragulator) | | Descriptor: | 9-{5-O-[(S)-hydroxy{[(R)-hydroxy(thiophosphonooxy)phosphoryl]oxy}phosphoryl]-alpha-L-arabinofuranosyl}-3,9-dihydro-1H-purine-2,6-dione, Folliculin, Folliculin-interacting protein 2, ... | | Authors: | Fromm, S.A, Young, L.N, Hurley, J.H. | | Deposit date: | 2019-02-13 | | Release date: | 2019-11-06 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural mechanism of a Rag GTPase activation checkpoint by the lysosomal folliculin complex.
Science, 366, 2019
|
|
5HM9
 
 | | Crystal structure of MamO protease domain from Magnetospirillum magneticum (apo form) | | Descriptor: | MamO protease domain, poly(UNK) | | Authors: | Hershey, D.M, Ren, X, Hurley, J.H, Komeili, A. | | Deposit date: | 2016-01-15 | | Release date: | 2016-03-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | MamO Is a Repurposed Serine Protease that Promotes Magnetite Biomineralization through Direct Transition Metal Binding in Magnetotactic Bacteria.
Plos Biol., 14, 2016
|
|
5HMA
 
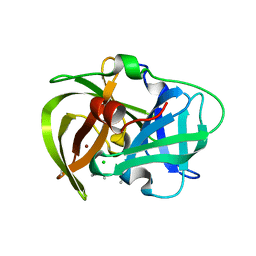 | | Crystal structure of MamO protease domain from Magnetospirillum magneticum (Ni bound form) | | Descriptor: | CHLORIDE ION, NICKEL (II) ION, Trypsin-like serine protease, ... | | Authors: | Hershey, D.M, Ren, X, Hurley, J.H, Komeili, A. | | Deposit date: | 2016-01-15 | | Release date: | 2016-02-10 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.299 Å) | | Cite: | MamO Is a Repurposed Serine Protease that Promotes Magnetite Biomineralization through Direct Transition Metal Binding in Magnetotactic Bacteria.
Plos Biol., 14, 2016
|
|
2F6M
 
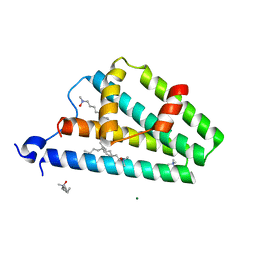 | | Structure of a Vps23-C:Vps28-N subcomplex | | Descriptor: | DECYLAMINE-N,N-DIMETHYL-N-OXIDE, MAGNESIUM ION, Suppressor protein STP22 of temperature-sensitive alpha-factor receptor and arginine permease, ... | | Authors: | Kostelansky, M.S, Lee, S, Kim, J, Hurley, J.H. | | Deposit date: | 2005-11-29 | | Release date: | 2006-04-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural and functional organization of the ESCRT-I trafficking complex.
Cell(Cambridge,Mass.), 125, 2006
|
|
2FAU
 
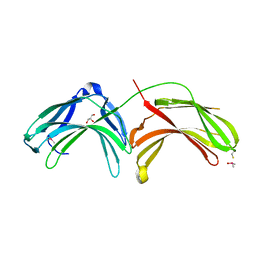 | | Crystal structure of human vps26 | | Descriptor: | GLYCEROL, Vacuolar protein sorting 26 | | Authors: | Shi, H, Rojas, R, Bonifacino, J.S, Hurley, J.H. | | Deposit date: | 2005-12-08 | | Release date: | 2006-05-30 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The retromer subunit Vps26 has an arrestin fold and binds Vps35 through its C-terminal domain.
Nat.Struct.Mol.Biol., 13, 2006
|
|
2FID
 
 | |
2F66
 
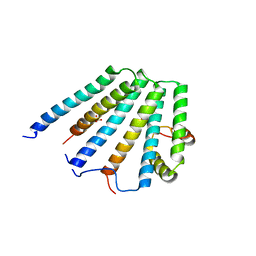 | | Structure of the ESCRT-I endosomal trafficking complex | | Descriptor: | Protein SRN2, SULFATE ION, Suppressor protein STP22 of temperature-sensitive alpha-factor receptor and arginine permease, ... | | Authors: | Kostelansky, M.S, Lee, S, Kim, J, Hurley, J.H. | | Deposit date: | 2005-11-28 | | Release date: | 2006-04-18 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural and functional organization of the ESCRT-I trafficking complex.
Cell(Cambridge,Mass.), 125, 2006
|
|
2FIF
 
 | |
3HTU
 
 | | Crystal structure of the human VPS25-VPS20 subcomplex | | Descriptor: | Vacuolar protein-sorting-associated protein 20, Vacuolar protein-sorting-associated protein 25 | | Authors: | Im, Y.J, Hurley, J.H. | | Deposit date: | 2009-06-12 | | Release date: | 2009-08-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure and function of the ESCRT-II-III interface in multivesicular body biogenesis.
Dev.Cell, 17, 2009
|
|
5C50
 
 | |
2PJW
 
 | |
4FZS
 
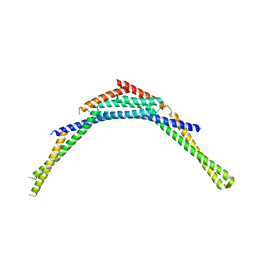 | | Structure of human SNX1 BAR domain | | Descriptor: | Sorting nexin-1 | | Authors: | van Weering, J.R.T, Sessions, R.B, Traer, C.J, Kloer, D.P, Bhatia, V.K, Stamou, D, Hurley, J.H, Cullen, P.J. | | Deposit date: | 2012-07-07 | | Release date: | 2013-07-10 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Molecular insight into vesicle-to-tubule membrane remodeling by SNX-BAR proteins
To be Published
|
|
4HMY
 
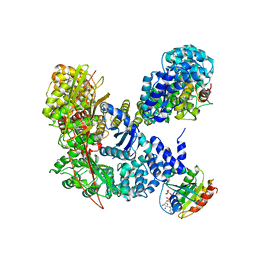 | | Structural basis for recruitment and activation of the AP-1 clathrin adaptor complex by Arf1 | | Descriptor: | ADP-ribosylation factor 1, AP-1 complex subunit beta-1, AP-1 complex subunit gamma-1, ... | | Authors: | Ren, X, Farias, G.G, Canagarajah, B.J, Bonifacino, J.S, Hurley, J.H. | | Deposit date: | 2012-10-18 | | Release date: | 2013-03-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (7 Å) | | Cite: | Structural Basis for Recruitment and Activation of the AP-1 Clathrin Adaptor Complex by Arf1.
Cell(Cambridge,Mass.), 152, 2013
|
|
4HPQ
 
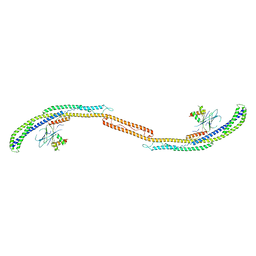 | | Crystal Structure of the Atg17-Atg31-Atg29 Complex | | Descriptor: | Atg17, Atg29, Atg31 | | Authors: | Stanley, R.E, Ragusa, M.J, Hurley, J.H. | | Deposit date: | 2012-10-24 | | Release date: | 2012-12-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.06 Å) | | Cite: | Architecture of the atg17 complex as a scaffold for autophagosome biogenesis.
Cell(Cambridge,Mass.), 151, 2012
|
|
4IKN
 
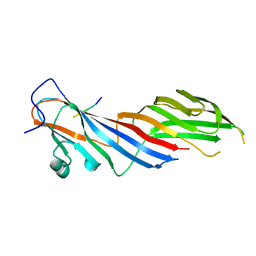 | | Crystal structure of adaptor protein complex 3 (AP-3) mu3A subunit C-terminal domain, in complex with a sorting peptide from TGN38 | | Descriptor: | AP-3 complex subunit mu-1, Trans-Golgi network integral membrane protein TGN38 | | Authors: | Mardones, G.A, Kloer, D.P, Burgos, P.V, Bonifacino, J.S, Hurley, J.H. | | Deposit date: | 2012-12-26 | | Release date: | 2013-02-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.851 Å) | | Cite: | Structural basis for the recognition of tyrosine-based sorting signals by the mu 3A subunit of the AP-3 adaptor complex.
J.Biol.Chem., 288, 2013
|
|
3MHV
 
 | | Crystal Structure of Vps4 and Vta1 | | Descriptor: | Vacuolar protein sorting-associated protein 4, Vacuolar protein sorting-associated protein VTA1 | | Authors: | Yang, D, Hurley, J.H. | | Deposit date: | 2010-04-09 | | Release date: | 2010-10-06 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural role of the Vps4-Vta1 interface in ESCRT-III recycling
Structure, 18, 2010
|
|
3OBU
 
 | |
3OBQ
 
 | |
